The best predictive model for hepatocellular carcinoma in patients with chronic hepatitis B infection
-
Jung Hwan Yu1, Soon Gu Cho2, Young-Joo Jin1, Jin-Woo Lee1

- Received August 30, 2021 Revised November 10, 2021 Accepted November 25, 2021
- ABSTRACT
-
Chronic hepatitis B (CHB) seriously threatens human health. About 820,000 deaths annually are due to related complications such as hepatitis B and hepatocellular carcinoma (HCC). Recently, the use of oral antiviral agents has significantly improved the prognosis of patients with CHB infection and reduced the risk of HCC. However, hepatitis B virus still remains a major factor in the development of HCC, raising many concerns. Therefore, numerous studies have been conducted to assess the risk of HCC in patients with CHB infection and many models have been proposed to predict the risk of developing HCC. However, as each study has different models for predicting HCC development that can be applied depending on the use of antiviral agents or the type of antiviral agents, it is necessary to properly understand characteristics of each model when using it for the evaluation of HCC in patients with CHB infection. In addition, because different variables such as host factor, viral activity, and cirrhosis are used to evaluate the risk of HCC development, it is necessary to assess the risk by carefully verifying which variables are used. Recently, studies have also evaluated the risk of HCC using risk prediction models through transient elastography and artificial intelligence (AI) system. These HCC risk predication models are also noteworthy. In this review, we aimed to compare HCC risk prediction models in patients with CHB infection reported to date to confirm variables used and specificity between each model to determine an appropriate HCC risk prediction method.
- INTRODUCTION
- INTRODUCTION
Hepatitis B virus (HBV) is the major cause of liver cirrhosis and hepatocellular carcinoma (HCC) and has infected more than 200 million people worldwide [1,2]. Efforts to predict the incidence of HCC in patients with chronic hepatitis B (CHB) infection have continued as HCC is an important factor in determining the prognosis [3,4]. It is known that the use of oral antiviral agents for HBV reduces the risk of HCC development in patients with CHB infection [5]. However, HCC risk is not completely eliminated despite the use of highly active antiviral agents such as entecavir (ETV) or tenofovir disoproxil fumarate (TDF) [6-10]. Therefore, efforts to evaluate the residual risk of HCC in patients taking antiviral drugs are continuously made, and various models for this purpose are continuously introduced.Antiviral therapy for CHB infection, especially in patients receiving potent antiviral agents such as ETV and TDF, may achieve extremely low or undetectable HBV DNA levels and normalized serum alanine aminotransferase (ALT) levels [11-13]. Therefore, it may be inappropriate to use factors indicative of viral activity, such as HBV DNA level, HBeAg, and ALT, as variables for the prediction of HCC in these patients. In practice, most studies of patients with CHB infection taking antiviral drugs do not take advantage of these factors and propose an HCC prediction model. However, as there are also many patients with CHB infection who do not use antiviral drugs, the predictive HCC risk in untreated patients is also important. Therefore, it is necessary to classify an HCC prediction model according to whether the patients take antiviral drugs.Regardless of the use of antiviral drugs, the most important predictor of HCC is the presence or absence of cirrhosis. Similarly, cirrhosis is an important factor in almost all HCC prediction models, and the highest weight is given to HCC prediction models evaluated by an integer scoring system. Therefore, it is important to accurately evaluate the presence or absence of cirrhosis in patients with CHB infection. In clinical practice, imaging tests and clinical diagnosis are used instead of histological evaluation through liver biopsy in most cases. To compensate for this, models using the FibroScan® (EchoSens, Paris, France) test, which can indirectly predict cirrhosis by measuring liver stiffness, have been introduced as a variable for predicting HCC development, but more validation studies are still needed to confirm the usefulness of these models.We also reported a purified and simple predictive model that can predict the occurrence of HCC in patients taking antiviral drugs. Although it showed a high predictive ability for HCC development, there were limitations in the variables that could be used depending on whether antiviral drugs were administered or not, and there was a disadvantage that the verification was not sufficient [14]. Through this review, we tried to identify HCC prediction models for patients with CHB infection reported to date and to verify the variables used in each study. In addition, we compared the differences in HCC prediction models according to whether antiviral drugs were used, and examined the most suitable HCC prediction model in patients with CHB infection.
- VARIABLES USED TO PREDICT HCC RISK
- VARIABLES USED TO PREDICT HCC RISK
Various factors related to the host, viral activity, and cirrhosis are used to predict the occurrence of HCC. Typically, the host factors used to predict HCC are age and sex while there are also other models that use family histories of HCC or diabetes as variables. However, most studies have used only age or the combination of age and sex as host factors. Variables related to viral activity, such as HBeAg, HBV DNA, and elevated ALT levels, have been recognized as useful factors for predicting the occurrence of HCC and have been used in many HCC prediction models. However, recently, in patients using antiviral drugs, the factors related to viral activity were not normalized or detected; therefore, their significance as a predictor of HCC in these patients, diminished. Thus, most of the studies conducted on patients using antiviral drugs, especially potent antiviral agents such as ETV and TDF, have not utilized these factors to predict HCC.Cirrhosis is present in 70–80% of patients with HCC, and the presence or absence of cirrhosis is considered an important variable in a predictive model of HCC. However, the diagnosis of liver cirrhosis in the clinical practice is sometimes unclear because it is diagnosed using imaging tests and clinical symptoms rather than using pathological results. In addition, since it is not easy to use the degree of liver fibrosis as a variable, it limits the ability to predict the risk of HCC based only on the presence or absence of cirrhosis. To compensate for this, the liver stiffness measurement (LSM) value is sometimes used as a variable in a HCC occurrence prediction model. However, given the additional testing cost, more validation is needed to determine its effectiveness as a variable in HCC predictive models.In addition to the previously known variables related to the occurrence of HCC, studies on new factors related to the occurrence of HCC in patients with CHB continue to be conducted. Recently, a study result indicated that the serum N-glycan biosignatures could be useful in the early diagnosis of HCC [15]. In addition, there have been studies showing that the gamma glutamyl transferase (γGT) isoenzyme II, cartilage oligomeric matrix protein (COMP), interleukin (IL)-6, serum programed death receptor (sPD) 1, and HBV pre-S mutants are also helpful in predicting the development of HCC [16]. In each of these studies, the factors related closely to the risk of HCC in CHB patients have the potential to be used as a variable in a predictive model for HCC (Fig. 1). However, analyses and verification of the predictive power of these factors and HCC models have not yet been conducted. It is recommended that future research focus on these areas.
- HCC RISK MODELS IN UNTREATED PATIENTS WITH CHB INFECTION
- HCC RISK MODELS IN UNTREATED PATIENTS WITH CHB INFECTION
Studies on HCC prediction models for patients who have not used antiviral drugs have mainly been conducted in Asia. The GAG-HCC (guide with age, gender, HBV DNA, core promoter mutations, and cirrhosis) score was developed from a cohort of 820 Chinese patients from tertiary referral clinics with CHB infections [17]. All the patients were treatment-naïve at baseline and followed-up for a median of 6.4 years. Based on a cut-off value of 100, the sensitivity and specificity were 84.1% and 76.2% for the 5-year prediction of HCC, and 88.0% and 78.7% for the 10-year prediction, respectively. The Chinese University (CU)-HCC score was first derived using a cohort of 1,005 patients with CHB infections in Hong Kong and validated in an independent cohort of 424 Chinese patients with CHB infections [18].The CU-HCC score is composed of five factors (age, albumin, bilirubin, HBV DNA, and cirrhosis), and divided HCC risk into three categories (low, <5; medium, 5–19; high, ≥20). The 5-year prediction of HCC development was 98.3%, 90.5%, and 78.9% in the low-, medium-, and high-risk groups, respectively. In the low-risk patient group (score <5), the negative predictive value excluding future HCC was 98.3–100.0%. The REACH-B score (risk estimation for HCC in CHB) was derived using 3,584 Chinese patients with CHB infections from the Taiwanese REVEAL cohort and validated in a cohort of 1,505 patients in Hong Kong and Korea [19]. The variables included in this risk score were sex, age, HBeAg, and ALT and HBV DNA levels. The risk of HCC development was simplified into an integer scoring system, with a range of 0 to 17 points. The risk of HCC occurrence ranged from 0.0% to 23.6% at 3 years, 0.0% to 47.4% at 5 years, and 0.0% to 81.6% at 10 years for patients with the lowest and highest risk of HCC, respectively. A revised version of the REACH-B score, the REACB-B II score, which includes quantitative serum HBsAg levels and HBV genotypes, was also studied [20].In addition, liver stiffness (LS) model, LSM-HCC, LSPS (LSM-spleen diameter to platelet ratio score), RWS (real-world risk)-HCC, AGED (age, gender HBeAg, and HBV DNA levels) scores, and the D2 AS model have been reported, indicating high area under the receiver operating characteristic (AUROC) values (Fig. 2) [21-26]. This shows that we can use various models to predict the occurrence of HCC in patients with CHB infections who have not received antiviral treatment (Table 1).
- HCC RISK MODELS IN CHB PATIENT TREATED WITH NUCLEOS(T)IDE ANALOGUES
- HCC RISK MODELS IN CHB PATIENT TREATED WITH NUCLEOS(T)IDE ANALOGUES
Since the recent AASLD, EASL, and APASL guidelines recommend ETV and TDF as the first-line treatment, most patients with CHB infections use these two drugs instead of the previously used lamivudine, adefovir, and telbivudine. Therefore, in the HCC risk prediction model for patients with CHB infections using antiviral drugs, many of the studies were conducted on patients who used the first-line drugs (ETV or TDF) while some studies were conducted on patients who only used ETV. The PAGE B study included 1,815 patients with CHB infections who received ETV or TDF for more than one year and was conducted on Caucasians, in contrast to previous studies that were conducted on Asians [27]. The PAGE B score used the variables of age, gender, and platelets, and the risk was divided into low (≤9), medium (10–17), and high (≥18). The 5-year cumulative HCC incidence in patients with CHB infections classified as low, medium, and high risk according to the PAGE-B score, were 0%, 3%, and 17%, respectively. In the validation cohort, the negative predictive value of HCC within 5 years approached 100%, based on a 10-point cut-off. Additionally, a modified PAGE B model in which 3,001 Koreans (including a validation cohort of 1,000) were studied with the addition of serum albumin to the PAGE B model, as a variable [28]. This model may also be used to predict the occurrence of HCC in patients with CHB infections, using either ETV or TDF.The CAMD (cirrhosis, age, male sex, and diabetes mellitus) model was developed in a study that was conducted in Hong Kong and Taiwan with 23,851 patients with CHB infections who were receiving either ETV or TDF [29]. Unlike other HCC risk prediction models, the CAMD score is characterized by the presence or absence of diabetes as a variable. In this study, two cut-off points, 8 and 13 points, were set to stratify patients into low-, medium-, or high-risk subgroups. The 3-year cumulative incidences of HCC in patients with a CAMD score of <8, 8–13, and >13 points were 0.27% (95% confidence interval [CI], 0.12–0.42%), 2.40% (95% CI, 2.03–2.78%), and 10.75% (95% CI, 9.68–11.81%), respectively. Another study that developed the AASL (age, albumin, sex, cirrhosis) model was conducted with 1,243 Korean patients with CHB infections receiving both ETV and TDF [14]. The AASL model classified the risk of HCC into low (≤5), intermediate (6–19), and high (≥20) risk groups with the 10-year cumulative HCC incidence rate being almost zero in the low-risk group. The 5-year cumulative incidences of HCC in the low-, intermediate-, and high-risk groups were 0%, 4.2%, and 17.6%, respectively. Among these models, the HCC-RESCUE and APA-B models were developed from studies that were conducted only on patients who were treated with ETV while the remaining models, the PAGE-B, CAMD, and AASL models were developed from studies on patients who were treated with either ETV or TDF. In addition to these models, studies that have been conducted in Asia have developed the HCC-RESCURE (risk estimating score in CHB patients using ETV), mREACH-B, PAGE-B-LS and the APA-B models (Table 2) [17,21-23].
- LSM IN HCC PREDICTION MODEL
- LSM IN HCC PREDICTION MODEL
As the degree of liver fibrosis is known to be related to the risk of HCC, it can be used as a useful factor in predicting the risk of HCC. However, a liver biopsy, the gold standard for evaluating the degree of liver fibrosis, is difficult to use in all patients with CHB infections in actual clinical practice because of its invasiveness and complications. Therefore, non-invasive transient elastography (FibroScan®, EchoSens) has been used to estimate liver stiffness, and its use in several HCC risk prediction models have been reported.Studies using LSM to predict HCC risk for patients with CHB infections developed the LS, LSM-HCC, and LSPS models. The LS model was developed in a study that was conducted using 1,250 patients with CHB infections in Korea with age, gender, HBV DNA, and LSM being used as variables to predict the development of HCC. Using these four variables, the AUROC of the HCC prediction model was 0.806 (95% CI, 0.738–0.874), a suitable value for the HCC risk prediction model. The LSM-HCC model was developed in a study that was conducted in Hong Kong using 1,555 patients with CHB infections and in which the risk was classified on a scale of 0 to 30 using LSM, age, albumin, and HBV DNA. In this study, a comparison of the AUROC values of LSM-HCC and CU-HCC indicated that the AUROC value of LSM-HCC was higher than that of CU-HCC (0.83–0.89 vs. 0.75–0.81). In addition, by applying a cut-off value of 11, the score excluded future HCC with high negative predictive values (99.4–100.0%) at 5 years. The LSPS model was used in a study on Korean patients with CHB infections in which the spleen diameter was used as a variable for predicting HCC. Although attempts were made to reflect portal hypertension-related cirrhosis complications in the prediction of HCC using the spleen diameter, the number of patients in this study was only 227, which was relatively small compared to the number of patients in the other models, and had the disadvantage of the spleen diameter not being a validated variable.
- VALIDATION OF HCC RISK MODELS
- VALIDATION OF HCC RISK MODELS
As models for predicting the risk of HCC in patients with CHB infections are continuously being published, studies to validate these models and compare them to find a more suitable model have also been reported. However, because the race, basic characteristics, and the types of antiviral drugs used in patients with CHB infections used to validate these models, may differ from that of previous studies, caution is required in their interpretation. In a 10-year follow-up study of 1,241 Korean patients with CHB, Seo et al. [34] compared and analyzed the PAGE-B, REACH-B, mREACH-B, and LSM-HCC models. According to this study, the mREACH-B score showed higher performance in predicting HCC than the PAGE-B and LSM-HCC groups at 3 years (AUC: 0.824 vs. 0.715 and 0.809, respectively), 5 years (AUC: 0.750 vs. 0.719 and 0.742, respectively), and 7 years (AUC: 0.770 vs. 0.714 and 0.765, respectively). A study that analyzed 14 liver cancer predictive models comparative, validated the REACH-B, CU-HCC, GAGHCC, PAGE-B, and mPAGE-B models in 986 Chinese patients with CHB infections using ETV [35]. According to this study, the HCC predictive power was generally acceptable for all models, with pooled AUCs ranging from 0.68 to 0.81 for 5-year, with the REAL-B score showing the highest discrimination (0.75 for 5-year prediction) and calibration (3-year Brier score 0.066) [36]. In a study comparing the AASL, RESCUE-B, PAGE-B, and mPAGE-B scores in 3,171 patients with CHB infections receiving both ETV and TDF, the predictive accuracy of the AASL score was the highest for the 3- and 5-year HCC predictions (AUC: 0.818 and 0.816, respectively), followed by RESCUE-B, PAGE-B, and mPAGE-B scores (AUC: 0.780–0.815 and 0.769–0.814, respectively) [37]. However, since this study was validated in the same area (Korea) where the AASL model was developed, there may be limitations in accepting the results as they are. Therefore, a validation study conducted on a large worldwide cohort is needed, in the future.
- HCC RISK PREDICTION MODEL USING ARTIFICIAL INTELLIGENCE (AI)
- HCC RISK PREDICTION MODEL USING ARTIFICIAL INTELLIGENCE (AI)
Currently, AI, including machine learning and deep learning, is being used widely in medical research and practice, and in the field of liver disease. HCC risk prediction aims to utilize AI for prognosis, diagnosis, and treatment purposes. Recently, a model for predicting HCC using AI was reported. It targeted to 6,051 Korean CHB patients, and an optimal model was constructed using the gradient-boosting machine method. In this study, the validation was performed on Koreans and Caucasians, and it was found that the predictive power of HCC was better than that of previously reported models such as the PAGE-B, REACH-B, and CU-HCC models [38]. Therefore, it is thought that in the future, a HCC occurrence prediction model using AI will be developed and studied, and as suggested by this study that such a HCC occurrence prediction model will become more precise. However, research on AI is still in its infancy, and more studies with be required to overcome any AI errors.
- IDEAL HCC RISK PREDICTION MODEL FOR CHB PATIENTS
- IDEAL HCC RISK PREDICTION MODEL FOR CHB PATIENTS
As the use of antiviral agents affects factors is related to viral activity, such as HBV DNA, qHBsAg, HBeAg, and ALT levels, different strategies are required depending on whether antiviral agents are used when developing an HCC predictive model for patients with CHB infections. However, if an HCC prediction model is developed according to whether antiviral drugs are administered to patients with CHB infections, it can be disadvantageous in terms of utilization. Therefore, if there is no significant difference in the predictive power of HCC, excluding the variables related to viral activity will be beneficial in the construction of a model predicting the occurrence of HCC that can be used regardless of the use of antiviral drugs. While it is advantageous to use an accurate calculation formula that takes into account the weight of the risk to predict the risk of HCC accurately, it is disadvantageous to use in actual clinical practice. For this reason, models such as the REACH B, PAGB B, and AASL models classify the risk of HCC using a simple integer score, which has provided satisfactory results with regard to the predictive power of HCC (Table 3). Therefore, the use of a simple scoring system rather than a complex formula appears to be useful for developing a widely accepted predictive model for HCC in patients with CHB infections (Fig. 3). Finally, if the inclusion of the recently highlighted AI, including machine learning and deep learning, is used appropriately, a more suitable liver cancer occurrence prediction model may be developed.
- FOOTNOTES
- FOOTNOTES
-
Authors’ contributions JU Yu wrote the manuscript and prepared the figures and tables. SG Cho, YJ Jin and JW Lee revised the manuscript. All the authors read and approved the final version.
Conflicts of Interest The authors have no conflicts to disclose.
Figure 1.
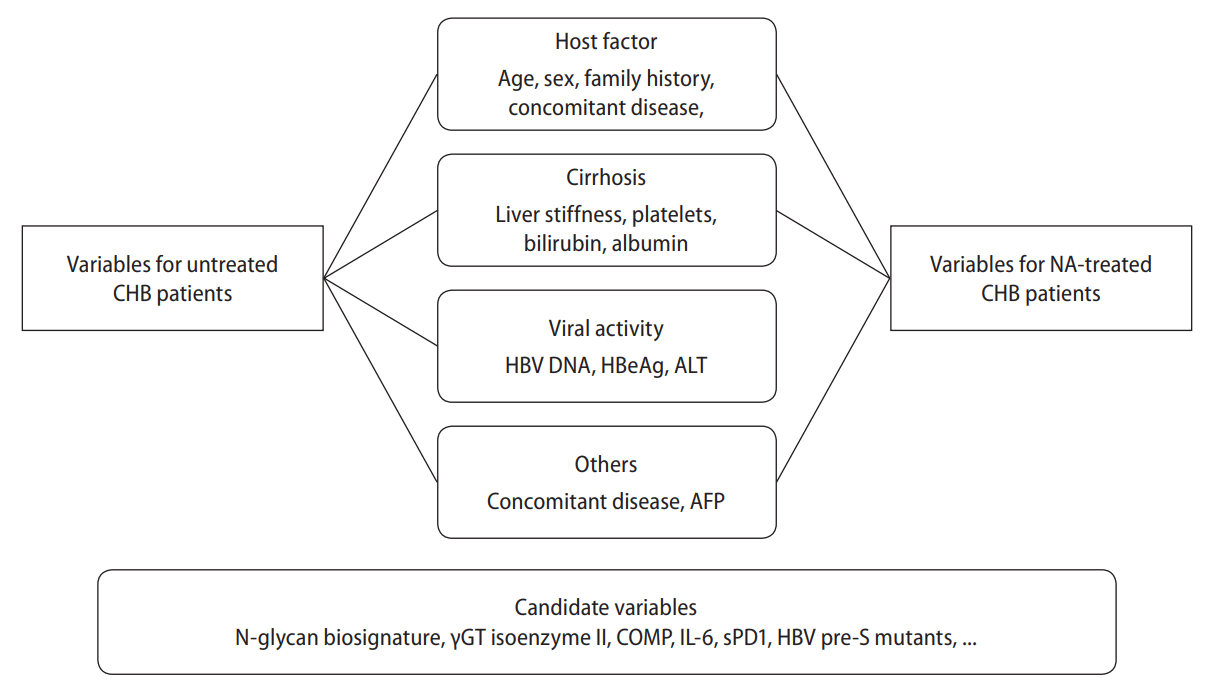
Figure 2.

Figure 3.

Table 1.
| HCC risk model | Country or area | Patients |
Variables used in the HCC risk prediction |
Number of variables | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Age | Sex | HBV-DNA | Cirrhosis | LSM | Others | |||||
| GAG-HCC | Hong Kong | 820 | √ | √ | √ | √ | 4 | [17] | ||
| CU-HCC | Hong Kong | 1,055 | √ | √ | √ | Albumin, bilirubin | 5 | [18] | ||
| REACH-B | Asia | 3,584 | √ | √ | √ | HBeAg, ALT | 5 | [19] | ||
| REACH-B II | Taiwan | 2,227 | √ | √ | √ | HBeAg, ALT, qHBsAg, genotype, family history | 8 | [20] | ||
| LS model | Korea | 1,250 | √ | √ | √ | √ | 4 | [21] | ||
| LSM-HCC | Hong Kong | 1,035 | √ | √ | √ | Albumin | 4 | [22] | ||
| LSPS | Korea | 227 | √ | PLT, spleen size | 3 | [23] | ||||
| RWS-HCC | Singapore | 583 | √ | √ | √ | AFP | 4 | [24] | ||
| AGED | China | 628 | √ | √ | √ | HBeAg | 4 | [25] | ||
| D2AD | Korea | 971 | √ | √ | √ | 3 | [26] | |||
Table 2.
| HCC risk model | Country or area | Patients | Antiviral agent ETV/TDF/others |
Variables used in the HCC risk prediction |
Number of variables | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Age | Sex | Cirrhosis | LSM | Others | ||||||
| PAGE-B | Europe | 1,325 | ETV or TDF | √ | √ | PLT | 3 | [27] | ||
| mPAGE-B | Korea | 2,001 | ETV or TDF | √ | √ | Albumin, PLT | 4 | [28] | ||
| PAGE-B-LS | Korea | 1,211 | 754/457/- | √ | √ | √ | PLT | 4 | [32] | |
| mREACH-B | Korea | 192 | 192/-/- | √ | √ | √ | ALT | 4 | [33] | |
| HCC-RESCUE | Korea | 990 | 990/-/- | √ | √ | √ | 3 | [30] | ||
| APA-B | Taiwan | 883 | 883/-/- | √ | PLT | 3 | [31] | |||
| CAMD | Asia | 23,851 | 22,971/880/- | √ | √ | √ | DM | 4 | [29] | |
| AASL | Korea | 944 | 601/342/- | √ | √ | √ | Albumin | 4 | [6] | |
| REAL-B | USA and Asia | 5,365 | 3,683/593/1,089 | √ | √ | √ | DM, PLT, AFP, alcohol use | 7 | [36] | |
Table 3.
- Abbreviations
- Abbreviations
AI artificial intelligence
ALT alanine aminotransferase
AUROC area under the receiver operating characteristic
CHB chronic hepatitis B
CI confidence interval
COMP cartilage oligomeric matrix protein
ETV entecavir
HBV hepatitis B virus
HCC hepatocellular carcinoma
IL interleukin
LS liver stiffness
LSM LS measurement
LSPS LSM-spleen diameter to platelet ratio score
RWS real-world risk
sPD serum programed death receptor
TDF tenofovir disoproxil fumarate
γGT gamma glutamyl transferase
- REFERENCES
- REFERENCES
REFERENCES
1. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018;391:1301-1314.
[Article] [PubMed]2. Kulik L, El-Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology 2019;156:477-491.e1.
[Article] [PubMed] [PMC]3. Ioannou GN. Chronic hepatitis B infection: a global disease requiring global strategies. Hepatology 2013;58:839-843.
[Article] [PubMed]4. Schmit N, Nayagam S, Thursz MR, Hallett TB. The global burden of chronic hepatitis B virus infection: comparison of country-level prevalence estimates from four research groups. Int J Epidemiol 2021;50:560-569.
[Article] [PubMed] [PMC]5. Liaw YF, Sung JJ, Chow WC, Farrell G, Lee CZ, Yuen H, et al. Lamivudine for patients with chronic hepatitis B and advanced liver disease. N Engl J Med 2004;351:1521-1531.
[Article] [PubMed]6. European Association for the Study of the Liver. EASL 2017 clinical practice guidelines on the management of hepatitis B virus infection. J Hepatol 2017;67:370-398.
[Article] [PubMed]7. Korean Association for the Study of the Liver (KASL). KASL clinical practice guidelines for management of chronic hepatitis B. Clin Mol Hepatol 2019;25:93-159.
[Article] [PubMed] [PMC]8. Terrault NA, Lok ASF, McMahon BJ, Chang KM, Hwang JP, Jonas MM, et al. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology 2018;67:1560-1599.
[Article] [PubMed] [PMC]9. Lee SB, Jeong J, Park JH, Jung SW, Jeong ID, Bang SJ, et al. Low-level viremia and cirrhotic complications in patients with chronic hepatitis B according to adherence to entecavir. Clin Mol Hepatol 2020;26:364-375.
[Article] [PubMed] [PMC]10. Lau G, Yu ML, Wong G, Thompson A, Ghazinian H, Hou JL, et al. APASL clinical practice guideline on hepatitis B reactivation related to the use of immunosuppressive therapy. Hepatol Int 2021;15:1031-1048.
[Article] [PubMed] [PMC]11. Chang TT, Lai CL, Kew Yoon S, Lee SS, Coelho HS, Carrilho FJ, et al. Entecavir treatment for up to 5 years in patients with hepatitis B e antigen-positive chronic hepatitis B. Hepatology 2010;51:422-430.
[Article] [PubMed]12. Marcellin P, Gane E, Buti M, Afdhal N, Sievert W, Jacobson IM, et al. Regression of cirrhosis during treatment with tenofovir disoproxil fumarate for chronic hepatitis B: a 5-year open-label follow-up study. Lancet 2013;381:468-475.
[Article] [PubMed]13. Lee SW, Choi J, Kim SU, Lim YS. Entecavir versus tenofovir in patients with chronic hepatitis B: enemies or partners in the prevention of hepatocellular carcinoma. Clin Mol Hepatol 2021;27:402-412.
[Article] [PubMed] [PMC]14. Yu JH, Suh YJ, Jin YJ, Heo NY, Jang JW, You CR, et al. Prediction model for hepatocellular carcinoma risk in treatment-naive chronic hepatitis B patients receiving entecavir/tenofovir. Eur J Gastroenterol Hepatol 2019;31:865-872.
[Article] [PubMed]15. Cong M, Ou X, Huang J, Long J, Li T, Liu X, et al. A predictive model using N-glycan biosignatures for clinical diagnosis of early hepatocellular carcinoma related to hepatitis B virus. OMICS 2020;24:415-423.
[Article] [PubMed]16. Van Hees S, Michielsen P, Vanwolleghem T. Circulating predictive and diagnostic biomarkers for hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol 2016;22:8271-8282.
[Article] [PubMed] [PMC]17. Yuen MF, Tanaka Y, Fong DY, Fung J, Wong DK, Yuen JC, et al. Independent risk factors and predictive score for the development of hepatocellular carcinoma in chronic hepatitis B. J Hepatol 2009;50:80-88.
[Article] [PubMed]18. Wong VW, Chan SL, Mo F, Chan TC, Loong HH, Wong GL, et al. Clinical scoring system to predict hepatocellular carcinoma in chronic hepatitis B carriers. J Clin Oncol 2010;28:1660-1665.
[Article] [PubMed]19. Yang HI, Yuen MF, Chan HL, Han KH, Chen PJ, Kim DY, et al. Risk estimation for hepatocellular carcinoma in chronic hepatitis B (REACH-B): development and validation of a predictive score. Lancet Oncol 2011;12:568-574.
[Article] [PubMed]20. Lee MH, Yang HI, Liu J, Batrla-Utermann R, Jen CL, Iloeje UH, et al. Prediction models of long-term cirrhosis and hepatocellular carcinoma risk in chronic hepatitis B patients: risk scores integrating host and virus profiles. Hepatology 2013;58:546-554.
[Article] [PubMed]21. Kim DY, Song KJ, Kim SU, Yoo EJ, Park JY, Ahn SH, et al. Transient elastography-based risk estimation of hepatitis B virus-related occurrence of hepatocellular carcinoma: development and validation of a predictive model. Onco Targets Ther 2013;6:1463-1469.
[Article] [PubMed] [PMC]22. Wong GL, Chan HL, Wong CK, Leung C, Chan CY, Ho PP, et al. Liver stiffness-based optimization of hepatocellular carcinoma risk score in patients with chronic hepatitis B. J Hepatol 2014;60:339-345.
[Article] [PubMed]23. Shin SH, Kim SU, Park JY, Kim DY, Ahn SH, Han KH, et al. Liver stiffness-based model for prediction of hepatocellular carcinoma in chronic hepatitis B virus infection: comparison with histological fibrosis. Liver Int 2015;35:1054-1062.
[Article] [PubMed]24. Poh Z, Shen L, Yang HI, Seto WK, Wong VW, Lin CY, et al. Real-world risk score for hepatocellular carcinoma (RWS-HCC): a clinically practical risk predictor for HCC in chronic hepatitis B. Gut 2016;65:887-888.
[Article] [PubMed]25. Fan C, Li M, Gan Y, Chen T, Sun Y, Lu J, et al. A simple AGED score for risk classification of primary liver cancer: development and validation with long-term prospective HBsAg-positive cohorts in Qidong, China. Gut 2019;68:948-949.
[Article] [PubMed]26. Sinn DH, Lee JH, Kim K, Ahn JH, Lee JH, Kim JH, et al. A novel model for predicting hepatocellular carcinoma development in patients with chronic hepatitis B and normal alanine aminotransferase levels. Gut Liver 2017;11:528-534.
[Article] [PubMed] [PMC]27. Papatheodoridis G, Dalekos G, Sypsa V, Yurdaydin C, Buti M, Goulis J, et al. PAGE-B predicts the risk of developing hepatocellular carcinoma in Caucasians with chronic hepatitis B on 5-year antiviral therapy. J Hepatol 2016;64:800-806.
[Article] [PubMed]28. Kim JH, Kim YD, Lee M, Jun BG, Kim TS, Suk KT, et al. Modified PAGE-B score predicts the risk of hepatocellular carcinoma in Asians with chronic hepatitis B on antiviral therapy. J Hepatol 2018;69:1066-1073.
[Article] [PubMed]29. Hsu YC, Yip TC, Ho HJ, Wong VW, Huang YT, El-Serag HB, et al. Development of a scoring system to predict hepatocellular carcinoma in Asians on antivirals for chronic hepatitis B. J Hepatol 2018;69:278-285.
[Article] [PubMed]30. Sohn W, Cho JY, Kim JH, Lee JI, Kim HJ, Woo MA, et al. Risk score model for the development of hepatocellular carcinoma in treatment-naïve patients receiving oral antiviral treatment for chronic hepatitis B. Clin Mol Hepatol 2017;23:170-178.
[Article] [PubMed] [PMC]31. Chen CH, Lee CM, Lai HC, Hu TH, Su WP, Lu SN, et al. Prediction model of hepatocellular carcinoma risk in Asian patients with chronic hepatitis B treated with entecavir. Oncotarget 2017;8:92431-92441.
[Article] [PubMed] [PMC]32. Chon HY, Lee HA, Suh SJ, Lee JI, Kim BS, Kim IH, et al. Addition of liver stiffness enhances the predictive accuracy of the PAGE-B model for hepatitis B-related hepatocellular carcinoma. Aliment Pharmacol Ther 2021;53:919-927.
[Article] [PubMed]33. Lee HW, Yoo EJ, Kim BK, Kim SU, Park JY, Kim DY, et al. Prediction of development of liver-related events by transient elastography in hepatitis B patients with complete virological response on antiviral therapy. Am J Gastroenterol 2014;109:1241-1249.
[Article] [PubMed]34. Seo YS, Jang BK, Um SH, Hwang JS, Han KH, Kim SG, et al. Validation of risk prediction models for the development of HBVrelated HCC: a retrospective multi-center 10-year follow-up cohort study. Oncotarget 2017;8:113213-113224.
[Article] [PubMed] [PMC]35. Wu S, Zeng N, Sun F, Zhou J, Wu X, Sun Y, et al. Hepatocellular carcinoma prediction models in chronic hepatitis B: a systematic review of 14 models and external validation. Clin Gastroenterol Hepatol 2021;19:2499-2513.
[Article] [PubMed]36. Yang HI, Yeh ML, Wong GL, Peng CY, Chen CH, Trinh HN, et al. Real-world effectiveness from the Asia Pacific Rim Liver Consortium for HBV risk score for the prediction of hepatocellular carcinoma in chronic hepatitis B patients treated with oral antiviral therapy. J Infect Dis 2020;221:389-399.
[Article] [PubMed]
