Endoplasmic reticulum stress and autophagy dysregulation in alcoholic and non-alcoholic liver diseases
Article information
Abstract
Alcoholic and non-alcoholic liver diseases begin from an imbalance in lipid metabolism in hepatocytes as the earliest response. Both liver diseases share common disease features and stages (i.e., steatosis, hepatitis, cirrhosis, and hepatocellular carcinoma). However, the two diseases have differential pathogenesis and clinical symptoms. Studies have elucidated the molecular basis underlying similarities and differences in the pathogenesis of the diseases; the factors contributing to the progression of liver diseases include depletion of sulfhydryl pools, enhanced levels of reactive oxygen and nitrogen intermediates, increased sensitivity of hepatocytes to toxic cytokines, mitochondrial dysfunction, and insulin resistance. Endoplasmic reticulum (ER) stress, which is caused by the accumulation of misfolded proteins and calcium depletion, contributes to the pathogenesis, often causing catastrophic cell death. Several studies have demonstrated a mechanism by which ER stress triggers liver disease progression. Autophagy is an evolutionarily conserved process that regulates organelle turnover and cellular energy balance through decomposing damaged organelles including mitochondria, misfolded proteins, and lipid droplets. Autophagy dysregulation also exacerbates liver diseases. Thus, autophagy-related molecules can be potential therapeutic targets for liver diseases. Since ER stress and autophagy are closely linked to each other, an understanding of the molecules, gene clusters, and networks engaged in these processes would be of help to find new remedies for alcoholic and non-alcoholic liver diseases. In this review, we summarize the recent findings and perspectives in the context of the molecular pathogenesis of the liver diseases.
INTRODUCTION
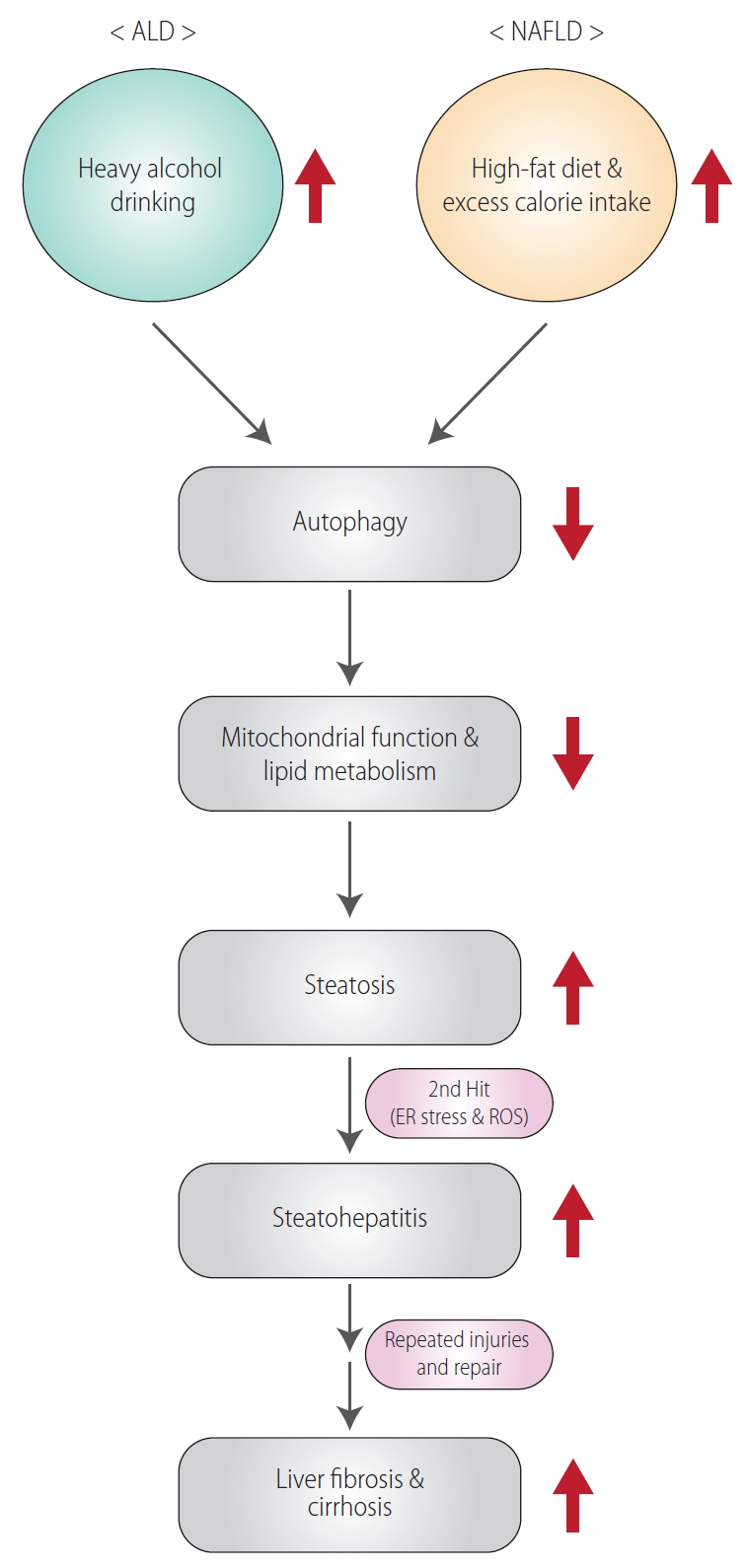
Excessive and chronic alcohol consumption may lead to the progression of alcoholic liver disease (ALD) [1]. ALD is caused by chronic alcohol consumption (>20 g/day for women and >30 g/day for men) [2,3]. Moreover, repeated alcohol drinking may promote progression of simple steatosis to steatohepatitis and/or cirrhosis [4]. Another most common liver disease is non-alcoholic fatty liver disease (NAFLD), which is considered as the hepatic manifestation of the metabolic syndrome including hypertension, type 2 diabetes, insulin-resistance, obesity, dyslipidemia, and distinct hepatic histological features [5]. Both ALD and NAFLD share general histopathological spectrum from simple steatosis to steatohepatitis and fibrosis, which may progress to more severe diseases (i.e., cirrhosis and hepatocellular carcinoma) (Fig. 1) [4]. However, the two diseases differ from each other in a variety of properties, ranging from the molecular mechanisms of disease exacerbation to differences in clinical features. In particular, infiltration of inflammatory cells occurs to a greater degree in ALD than in NAFLD. In contrast, fat degeneration in hepatocytes is more pronounced in NAFLD than in ALD [6]. Despite the ongoing study of the pathology, causes, and risk factors for the diseases, we do not yet have an appropriate treatment regimen. Here, we aim to motivate intensive research on the diseases by reviewing the current understanding of the causes of ALD and NAFLD, and the trends in potential therapeutic approaches.
GENERAL MOLECULAR PATHOGENESIS
Definition of endoplasmic reticulum (ER) stress
ER is a structure of a membrane component that extends from the nuclear membrane and has two types: a ribosome attached rough ER and a ribosome-free smooth ER. Approximately one-third of the protein in the cell is translated from the messenger RNA to protein, which becomes an active protein structure through processes such as folding, assembly, glycation, and disulfide bond [7]. Smooth ER is the site for the synthesis of lipids and sterols, and functions as a calcium reservoir to regulate the cellular calcium concentrations [8,9]. However, if either immature protein flows into the ER above the capacity, or calcium is depleted in the ER, the function of ER is impaired. This condition is defined as ER stress [9-11]. When ER stress occurs, cells have a defense mechanism to survive, which is called the ER stress response [8]. Thus, ER stress is a cellular stress, in which the levels of synthetic proteins exceed the capacity of unfolded protein responses. Finally, when the ER stress becomes so severe that it cannot be overcome and the ER cannot recover its function, then cell death pathway is activated with inflammatory responses [9,12].
Cross-links between ER and mitochondria
In several cell and animal models, we found ER stress as a key stimulus of fatty liver transition to non-alcoholic steatohepatitis (NASH). So, ER stress is claimed as a secondary hit to provoke hepatocyte injury and inflammation, leading to liver fibrosis in a chronic situation (Fig. 1). Dysregulation of unfolded protein responses enhances ER stress and consequently may promote catastrophic cell death [13]. Additionally, ER stress, in association with reactive oxygen species (ROS) production, triggers the subsequent activation of cell death pathway due to imbalance of redox homeostasis [14,15]. Cytochrome P450 2E1, a high affinity enzyme for ethanol metabolism, is located in the membrane of ER, and thus the induction of cytochrome P450 2E1 would be also associated with ER stress in hepatocytes inflicted by sustained ethanol consumption [16]. This idea would be also supported by impairment of free fatty acids (FFAs) oxidation in mitochondria because mitochondria and ER physiologically work together through mitochondria-associated ER membrane [17].
The lipids are mainly metabolized in the liver, and lipid metabolism in the liver maintains homeostasis by balancing or storing fat for energy [18]. In particular, lipid absorption, esterification, oxidation and secretion of fatty acids are performed in liver cells [19]. The supply of FFAs to the liver contributes to the pathogenesis of hepatic steatosis; triglycerides and cholesterol are stored in the form of lipid droplets to protect cells from exposure to excessive amounts of FFAs that can damage signal pathways and metabolic homeostasis [20]. Impairment of oxidative metabolism of FFAs is also coupled to fat accumulation because FFAs are used as fuel for mitochondrial oxidation.
Compared to normal individuals, the incidence of ALD increases in patients with fatty liver by 2–3 times [21]. Thus, obesity is considered as an independent risk factor for alcoholic steatohepatitis since diet pattern and dietary fat content contribute to the progression of ALD. Mitochondrial dysfunction occurs not only by peroxidation of unsaturated fats of mitochondrial phospholipids, but by the attack of oxygen free radicals generated through ethanol metabolism. Hence, excessive alcohol consumption causes impairment of mitochondrial function [22]. In NAFLD patients, dysregulation of fatty acid metabolism causes hepatic steatosis and hyperlipidemia in a large patient population. In addition, NAFLD as a result of obesity and diabetes facilitates inflammatory cytokines production [23]. So, metabolic profile in hepatocytes greatly alters because of fuel source and consumption rates change. Together, metabolic disturbances caused by fat along with alcohol consumption stimulate worsening of alcoholic steatohepatitis, which may lead to more severe conditions.
MITOCHONDRIAL TARGETS
Peroxisome proliferator-activated receptor alpha regulates the expression of lipid metabolism-related genes (e.g., Cyp4a1, Acbp, and Acsl) [24]. In our preliminary study, we found that mitochondrial activity and oxidative energy metabolism are controlled by core molecules including sirtuin 1, peroxisome proliferator-activated receptor-γ coactivator 1-alpha and peroxisome proliferator-activated receptor alpha (i.e., a transcription complex required for mitochondrial biogenesis) [25,26]. In this study, we have shown that an activated form of G protein subunit alpha 12 (Gα12) stabilizes sirtuin 1 via ubiquitin specific peptidase 22 induction, which depends on hypoxia-inducible factor 1-alpha. The research results showed that Gα12 regulates sirtuin 1-dependent lipid oxidation in mitochondria through hypoxia-inducible factor 1-alpha-mediated ubiquitin specific peptidase 22 induction, identifying Gα12 as a linker between cell surface signaling and mitochondrial lipid oxidation (Fig. 2).
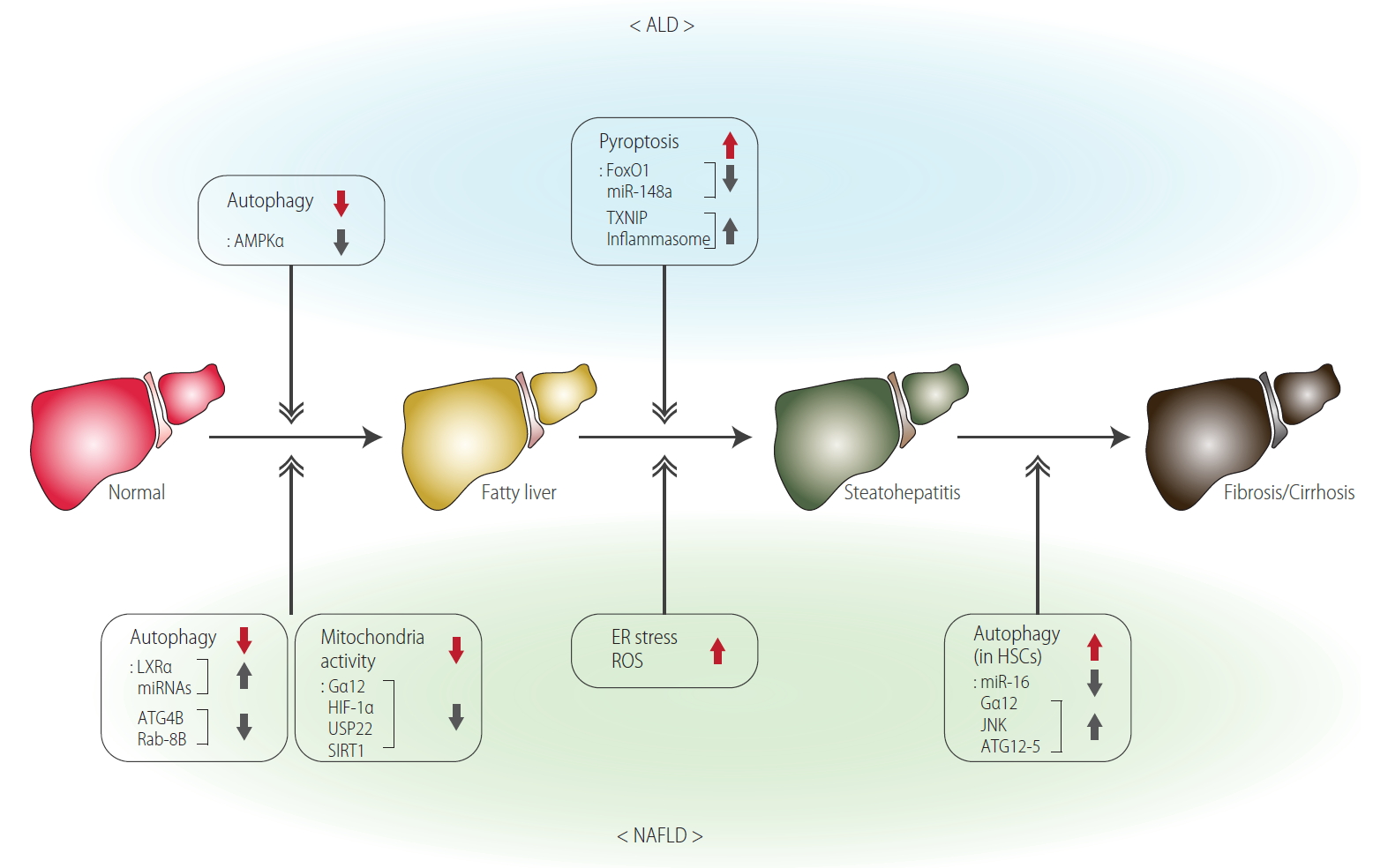
Functional molecular biomarkers for the processes of ALD and NAFLD. ALD, alcoholic liver disease; AMPKα, AMP-activated protein kinase alpha; miR, microRNA; TXNIP, thioredoxin interacting protein; LXRα, liver X receptor α; ATG4B, autophagy related 4B cysteine peptidase; Gα12, G protein subunit alpha 12; HIF-1α, hypoxia-inducible factor 1-alpha; USP22, ubiquitin specific peptidase 22; SIRT1, sirtuin 1; ER, endoplasmic reticulum; ROS, reactive oxygen species; HSCs, hepatic stellate cells; JNK, c-Jun N-terminal kinase; ATG12-5, autophagy related 12-5 conjugate; NAFLD, non-alcoholic fatty liver disease.
Liver fibrosis is manifested by repeated wound healing processes, such as an increase in matrix protein and a decrease in matrix remodeling: liver fibrosis is characterized by decreased matrix remodeling and increased production of matrix proteins, and may lead to end-stage cirrhosis [27]. Liver fibrosis is defined by concerted actions of non-parenchymal cells of the liver, particularly macrophages (including Kupffer cells), hepatic stellate cells (HSCs), and endothelial cells [28]. Accumulation of aberrant extracellular matrix is induced by a diverse population of myofibroblasts, among which HSCs play a major role [29].
AUTOPHAGY TARGETS
Definition of autophagy
Studies have reported the correlation between hepatic diseases and autophagy. Autophagy may inhibit the progression of steatosis and fatty hepatitis by preventing hepatocyte injury [30,31].
The liver is the major detoxifying and metabolic organ. Autophagy, a “recycling mechanism” in hepatocytes, is evolved to salvage key metabolites and to provide energy for sustaining anabolism [32]. Thus, autophagy is a crucial regulator of cellular homeostasis [32-34]; autophagy targets damaged cellular constituents, such as denatured proteins, accumulated fat, and mitochondria that have lost their function to lysosomes for degradation [35,36]. Amino acids, insulin, and mammalian target of rapamycin signaling pathways inhibit the autophagic pathway [36]. Additionally, fasting regulates metabolic pathways, such as gluconeogenesis, ketone body formation, and β-oxidation. Fatty acids are mainly generated by selective autophagy of lipid droplets (lipophagy), while amino acids are generated by proteolysis through autophagy and are used for gluconeogenesis [33,37]. Therefore, autophagy plays a decisive role in regulating liver physiology and balancing liver metabolism [38].
Autophagic molecules associated with liver steatosis
Chronic alcohol intake or excessive energy intake causes damage to the regulation of autophagy. When it persists, the homeostatic balance breaks and metabolic diseases develop [39,40]. Although several studies have demonstrated the correlation between autophagy and metabolic liver disease, the role of autophagy in metabolic liver disease has not been completely elucidated. Moreover, the understanding of the regulatory molecules by which the disease progression is determined is still needed.
The role of autophagy in acute and chronic ALD is still complex and controversial. It is generally known that binge drinking enhances autophagy of hepatocytes, which limits hepatic cell damage, death and fat accumulation by selectively removing excess lipid droplets and damaged mitochondria [41]. In contrast, chronic alcohol intake has been reported to decrease lysosome function and increase ubiquitinated aggregates accumulation [42]. Thus, the duration of alcohol exposure affects autophagy [43]. 5’ AMP-activated protein kinase, an enzyme that plays a role in the metabolism of fatty acids (e.g., acetyl-CoA carboxylase), is a key player of autophagy. Ethanol consumption causes inhibition of 5’ AMP-activated protein kinase, suggestive of the role of autophagy in ALD. One of the studies claims a mechanism for suppressing autophagy during chronic alcohol intake in association with decrease of 5’ AMP-activated protein kinase activity, which may increase fat production through decrease of β-oxidation [44]. This information can be employed to understand the pathogenesis of ALD.
Although the evaluation of autophagy in animal and human samples is somewhat controversial [45], it is generally agreed that autophagy is reduced in NAFLD. Hepatocellular steatosis is aggravated in mice deficient in autophagy-related genes [46,47]. Inhibition of hepatocyte-specific run domain beclin-1-interacting and cysteine-rich domain-containing protein (beclin-1 interacting negative regulator for autophagosome fusion), overexpression of adenoviral-induced autophagy-related 7 [47,48], or treatment of an autophagy activator such as rapamycin can relieve ER stress and hepatic steatosis. The opposite effect was seen with the inhibitor [49]. Moreover, autophagy participates in the basic conversion process of lipids by decomposing lipid droplets under physiological conditions. Recently, we demonstrated that liver X receptor α (LXRα) functions as a transcriptional regulator of microRNAs (miRs) [50]. In this study, gene set enrichment analysis was used to discover the phenomenon that autophagy can be inhibited by LXRα, and focusing on this, we proved the regulatory mechanism of autophagy using animal model. As a mechanistic basis, it was noted that LXRα inhibits autophagy-related 4B cysteine peptidase and rasrelated protein Rab-8B, and this effect is due to the transcriptional induction of let-7a and miR-34a by LXRα. In addition, the same mechanism works in human samples and genetic obesity-derived animal (ob/ob, db/db) samples. Furthermore, LXRα and the above identified molecules all inhibit lipid degradation and mitochondrial function, supportive of a link between autophagy and NAFLD when LXRα is activated. The outcomes may provide key information for target discovery and potential strategy to treat NASH (Fig. 2).
Autophagic molecules associated with liver fibrosis
Autophagy is reported to be involved in the progression of liver fibrosis [51]. Activation of HSCs is a major phenomenon in liver fibrosis because these cells differentiate into myofibroblasts and produce a major extracellular matrix in the liver [52]. HSCs of the quiescent phenotype store vitamin A in lipids and are activated when vitamin A is decomposed by various stimuli. Thus, HSCs can be activated by promoting autophagy. Previously, we have shown that Gα12 activation increases autophagy in HSCs, accompanying c-Jun N-terminal kinase-dependent autophagy-related 12-5 conjugation [53]. In the study, miR-16 directly inhibits de novo synthesis of Gα12 and thereby modulations of miR-16 alter autophagy (Fig. 2). Consistently, patients with severe fibrosis exhibited downregulated levels of miR-16 [53]. The activation of HSCs by apoptotic hepatocytes or by transforming growth factor-β produced from activated Kupffer cells also contributes to fibrosis [54]. In this process, sustained ER stress, cytokines, saturated FFAs, and adipokines could be also involved [54].
Potential targeting of autophagy
Currently, there are no specific treatments for non-alcohol/alcohol-related liver disease, and the main treatment is dietary control or alcohol withdrawal. Recent studies have demonstrated the importance of autophagy in metabolic diseases and suggested new treatment strategies. Preclinical studies have reported that enhancing autophagy using carbamazepine or rapamycin decreases fat accumulation and liver damage in obese and chronic ethanolfed mice [49]. Although there is a Janus face of autophagy in chronic liver disease, autophagy is primarily a hepatoprotective mechanism; autophagy is protective against the early stages of hepatocellular carcinoma and promotes liver regeneration [55]. Conversely, autophagy may be deleterious during the late phase of hepatocellular carcinoma in addition to activation of HSCs [51,56]. Thus, the cell-specific and pathology-specific regulation of autophagy should be considered for devising strategies for liver disease treatment.
CHANGES IN LIPID PROFILES
Alcohol-induced hepatic dysfunction or injury is closely related to abnormal lipid profiling [57,58]. Cumulated evidence shows that liver steatosis is linked to insulin resistance and the consequent hyperinsulinemia. In addition to steatosis, chronic alcohol consumption induces insulin resistance in the liver as indicated by the suppression of insulin-responsive genes [59,60]. Moreover, heavy alcohol consumption may lead to impairment of mitochondrial membrane depolarization; dysregulation of cellular redox state results at least in part from changes in the nicotinamide adenine dinucleotide (NAD+)/nicotinamide adenine dinucleotide hydride (NADH) ratio [61,62]. Alcohol disrupts the sirtuin 1 signaling pathway, which is involved in the regulation of mitochondrial biogenesis and oxygen consumption, and consequently promotes hepatic steatosis, injury, and inflammation [63]. In particular, mitochondrial dysfunction contributes to this change to a large extent, in which replacement of old non-functional mitochondria is disturbed due to autophagy inhibition as well as impaired proteasome function [64,65]. Electron microscopy has revealed the presence of megamitochondria in the liver of patients with ALD [66,67].
Lipotoxicity not only causes inflammation, ER stress, and ROS, but also affects the biological function of organelles, the most important of which is the mitochondria [68]. Increasing autophagy removes damaged mitochondria in a non-alcoholic fatty liver animal model, improving fatty liver [69]. Conversely, when autophagy is inhibited, severe mitochondrial damage and hepatic steatosis occur [70]. Additionally, suppression of autophagy through drug or gene regulation can reduce the number of triglycerides and lipid particles in hepatocytes [71]. As a result, a decrease in FFAs supply from lipid particles reduces mitochondrial β-oxidation, and the consequential fat accumulation in turn increases mitochondrial dysfunction [72].
Therefore, the interactions among mitochondrial activity, biogenesis, and autophagy contribute to metabolic dysregulation of hepatocytes in alcoholic and non-alcoholic liver diseases.
ALD AND DRUG CANDIDATES
Alcoholic liver steatosis is considered as a benign condition and is thus reversible to normal state under the condition of abstinence [73,74]. One of the early hallmarks of alcohol-induced liver damage caused by alcohol is the accumulation of lipid droplets in hepatocytes [75]. Clinically, most patients experience no symptoms for alcohol-induced fatty liver. However, patients with alcoholic steatohepatitis or fibrosis may present symptoms such as nausea and vomiting, which are caused by dysregulated cytokines production [76-79]. In a small proportion of chronic and heavy alcohol drinkers, ALD may lead to the development of liver cirrhosis (Fig. 1) [80-84].
Electrons generated during alcohol metabolism promote production of reactive intermediates, including ROS and reactive nitrogen species, which are catalyzed by enzymes such as alcohol dehydrogenase and cytochrome P450 2E1 [85,86]. ROS and reactive nitrogen species oxidize lipids to generate lipid peroxidation end products such as isoprostane and malondialdehyde [87-89]. Mitochondria is another organelle, from which ROS is greatly generated through oxidative fuel consumption [90]. Since an excess amount of ROS production causes depletion of reduced sulfhydryl pools (e.g., glutathione) in hepatocytes, hepatocytes may be sensitized by the stresses of toxic cytokines (e.g., tumor necrosis factor-β) and thus are more susceptible to injurious challenges. Previously, we had demonstrated that a spectrum of liver injury, including pyroptotic hepatocyte death, occurs in an alcohol abuse animal model; alcohol induces pyroptosis in hepatocytes by promoting ‘thioredoxin interacting protein and NOD-, LRR- and pyrin domain-containing protein 3’ inflammation through decreases of miR-148a and forkhead box protein O1 [91]. Attention was also paid to methionine and S-adenosylmethionine in the field of metabolic mechanism studies, the levels of which may be changed by alcohol consumption as part of the evidence of changes in cellular glutathione pool [92].
As mentioned above, various mechanisms are involved in ALD, and based on these findings, clinical treatment with new and emerging molecular targets for ALD treatment is ongoing (Table 1).
NAFLD AND DRUG CANDIDATES
Globally, NAFLD is the most common liver disease that encompasses diseases from inflamed steatosis to NASH, cirrhosis, and hepatocellular carcinoma [93]. In Asian and American populations, the prevalence of NAFLD is approximately 30% and the disease is often accompanied by type 2 diabetes and obesity [94]. Accurate diagnosis of NASH is essential because inflammation and/or fibrosis may determine the prognosis of NASH. Additionally, advanced conditions such as cirrhosis may require liver transplantation accordingly [95]. Currently, there are increased efforts to develop therapeutic strategies for NASH, which have no established treatment.
Previous studies have reported that multiple hits triggered NAFLD, which was recognized as a potential therapeutic goal. Several modulators of these targets and related pathways have been analyzed in clinical trials (phase II) or are currently in ongoing clinical trials (Table 1) [93]. The candidate drugs inhibit de novo lipogenesis or increase lipid export from the liver [94]. Additionally, liver or systemic insulin resistance is improved by diverse drugs or gastrointestinal hormones such as peroxisome proliferator-activated receptor γ agonists. Accumulation of excess lipids may facilitate simple steatosis progression to inflamed steatosis. This step usually provides effective targeting because inflammation generally precedes fibrosis. Currently, the inhibitors of apoptosis signal-regulating kinase 1, C-C chemokine receptor types 2/5, and chemokine receptors of C-C motif chemokine 5 and C-C motif chemokine 2 are under phase III clinical trials [96]. Several farnesoid X receptor agonists reduce gluconeogenesis and liver fat production, inhibit bile acid synthesis, improve peripheral insulin sensitivity, and thereby have a profound effect on various pathways for development of NAFLD [97,98]. Several farnesoid X receptor agonists have been tested in clinical trials (Table 1) [99]. Combination therapy may be an effective therapeutic strategy for NAFLD as it has a complex pathophysiology.
CONCLUSION
In this review, we attempted to summarize the molecular mechanisms underlying ALD and NAFLD. Both diseases are associated with unhealthy lifestyle habits, including increased consumption of alcohol and unhealthy diet. Extraordinary progress has been made in the understanding of the pathology of fatty liver disease so far, suggesting that several parallel hits are required to conquer the disease. Understanding the molecular mechanisms underlying ALD and NAFLD is important for medical professionals, especially physicians, general practitioners, hepatologists, and diabetologists. Because fatty liver disease has many clinical features, doctors should consider the patients in a multifaceted way. Identifying the various molecular mechanisms and finding therapeutic functions are key clinical prerequisites. In the future, the awareness of the disease progression and key targets would enable proper diagnosis, effective management, and treatment of comorbid diseases.
Notes
Authors’ contribution
YSK and SGK contributed to the literature review, and manuscript preparation.
Conflicts of Interest: The authors have no conflicts to disclose.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (NRF-2018R1A2A1A05078694) and in part by Dongguk University fund.
Abbreviations
ALD
alcoholic liver disease
ER
endoplasmic reticulum
FFAs
free fatty acids
Gα12
G protein subunit alpha 12
HSCs
hepatic stellate cells
LXRα
liver X receptor α
miR
microRNA
NAD
nicotinamide adenine dinucleotide
NADH
nicotinamide adenine dinucleotide hydride
NAFLD
non-alcoholic fatty liver disease
NASH
non-alcoholic steatohepatitis
ROS
reactive oxygen species