Performance evaluation of the HepB Typer-Entecavir kit for detection of entecavir resistance mutations in chronic hepatitis B
Article information
Abstract
Background/Aims
Molecular diagnostic methods have enabled the rapid diagnosis of drug-resistant mutations in hepatitis B virus (HBV) and have reduced both unnecessary therapeutic interventions and medical costs. In this study we evaluated the analytical and clinical performances of the HepB Typer-Entecavir kit (GeneMatrix, Korea) in detecting entecavir-resistance-associated mutations.
Methods
The HepB Typer-Entecavir kit was evaluated for its limit of detection, interference, cross-reactivity, and precision using HBV reference standards made by diluting high-titer viral stocks in HBV-negative human serum. The performance of the HepB Typer-Entecavir kit for detecting mutations related to entecavir resistance was compared with direct sequencing for 396 clinical samples from 108 patients.
Results
Using the reference standards, the detection limit of the HepB Typer-Entecavir kit was found to be as low as 500 copies/mL. No cross-reactivity was observed, and elevated levels of various interfering substances did not adversely affect its analytical performance. The precision test conducted by repetitive analysis of 2,400 replicates with reference standards at various concentrations showed 99.9% agreement (2398/2400). The overall concordance rate between the HepB Typer-Entecavir kit and direct sequencing assays in 396 clinical samples was 99.5%.
Conclusions
The HepB Typer-Entecavir kit showed high reliability and precision, and comparable sensitivity and specificity for detecting mutant virus populations in reference and clinical samples in comparison with direct sequencing. Therefore, this assay would be clinically useful in the diagnosis of entecavir-resistance-associated mutations in chronic hepatitis B.
INTRODUCTION
Despite the availability of highly effective and safe vaccines for more than 20 years, more than 400 million individuals worldwide are chronically infected with hepatitis B virus (HBV).1 Chronic hepatitis B (CHB) infection is associated with the development of cirrhosis, hepatocellular carcinoma (HCC), and death.2,3 Approximately 25% of patients with HBV will eventually die of liver failure and hepatocellular carcinoma if left untreated. In the REVEAL study, the most significant risk factor for the development of cirrhosis or HCC was directly proportional to serum HBV DNA levels,4-6 and it appears that the sustained suppression of serum HBV DNA replication is essential for impeding or reversing disease progression. Hence, an effective and prolonged suppression of HBV DNA, which reduces the risk of cirrhosis and HCC, is the primary treatment target. Antiviral therapy is used in CHB to minimize liver damage and limit disease progression.7
The roadmap concept was established by Keeffe et al.8 Early monitoring of virological responses to therapy in CHB treated with oral nucleos(t)ide analogs (NAs) is essential to identify primary treatment failure at week 12 and suboptimal responses at week 24 in order to modify the management accordingly. NAs can function as antiviral agents by inhibiting HBV replication and competing with the natural nucleotide substrate of DNA polymerase, thereby terminating the synthesis of viral DNA.9 However, it also often results in the emergence of drug resistant mutants and an ensuing treatment failure.
The deoxyguanosine nucleoside analogue entecavir (ETV) has been shown to be highly effective at suppressing HBV DNA replication to undetectable levels and normalizing alanine aminotransferase (ALT) in the treatment of patients with chronic hepatitis B,10-16 including patients with advanced fibrosis or cirrhosis and compensated liver disease.17,18 In vitro studies have shown that ETV is effective in suppressing adefovir (ADV) resistant mutants.19 ETV has been reported to be effective in suppressing HBV DNA levels in ADV resistant patients with prior lamivudine (LMV) resistance.20 ETV monotherapy may be considerably efficacious in cases with an initial virological response but its efficacy is attenuated by frequent emergence of ETV resistance in ADV refractory CHB patients with prior LMV resistance.21
The routine use of genotypic drug resistance testing requires convenient assays with high reproducibility that can be performed by laboratories skilled in molecular diagnostic techniques. The HepB Typer-Entecavir kit, which has recently acquired the Korea Food and Drug Safety (KFDA) approval for in vitro diagnostic use, is designed to provide mutant information of the codons 184, 202 and 250 in the polymerase (reverse transcriptase, rt) coding region by using restriction fragment mass polymorphism (RFMP) which is based on amplification and mass detection of oligonucleotides excised from type IIS enzyme digestion using matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry.
In order to determine the effects of specimen characteristics on the performance characteristics of the HepB Typer-Entecavir kit, a series of studies were undertaken to assess the effect of concentrations of HBV DNA, presence of interfering substances, and other viral nucleotides. Ability to detect drug resistance-associated mutations was also evaluated using a HBV DNA samples derived from molecular clones carrying a variety of resistance associated mutations in codons rt184, rt202 and rt250 and the assay performance was compared with results from direct sequencing assays.
MATERIALS AND METHODS
Patients
This study included 396 specimens stored in Yonsei Liver Blood Bank (YLBB) collected from 108 patients with chronic HBV infection who visited Severance Hospital, Seoul, Korea between February 2008 and June 2011.
Patients eligible for entry to the study were aged ≥20 years with serum HBV surface antigen (HBsAg) present ≥6 months and HBeAg-positive CHB being treated with LMV for ≥6 months. Eligible patients with partial virologic response or resistance against LMV receive ETV 0.5 mg or 1mg for ≥6 months, respectively. The upper limit of normal (ULN) for ALT was defined as 40 IU/L. Eligible patients had ALT <10×ULN with no evidence of HCC. All patients were tested for ETV mutations at baseline and at weeks 24, 48 and 72 in patients with serum HBV DNA≥60 IU/mL. Patients were excluded from this study if they had serological evidence of coinfection with HCV, HIV or hepatitis D virus. Those with decompensated liver disease, pregnant and breastfeeding women were also excluded. Experimental protocol followed the ethical guidelines of the 1975 Declaration of Helsinki, as reflected in a priori institutional review board approval. All the patients gave signed informed consent.
Serum specimens
In total, 396 serum specimens from 108 patients were taken and stored at -70℃ as 1 mL aliquots until use. Specimens were shipped on dry ice to the testing laboratories, where they were again stored at -70℃. HBV DNA was extracted from 200 µL of serum specimens using the QIAamp blood kit (Qiagen, Chatsworth, CA, USA) according to the manufacturer's instructions and dissolved in 20 µL of distilled water. 2 µL of the viral DNA was used for the PCR reaction, HBV DNA titer was quantified using COBAS AmpliPrep-COBAS TaqMan (detection limit: 20 IU/mL; Roche Diagnostics GmbH, Mannheim, Germany). 226 serum specimens were diagnosed with HBV DNA positive on the basis of HBV DNA titer (≥60 IU/mL). Among these specimens, 102 specimens had rt204M, and 124 specimens had rtM204I/V. Other 170 serum specimens were diagnosed with HBV DNA negative (HBV DNA <60 IU/mL). HBV negative human serum was purchased from a commercial source (Boston Biomedica, Inc., Cambridge, MA, USA).
Preparation of reference standards
Plasmids with type-specific inserts of the wild type and ETV resistance mutant viruses, which obtained from serum specimens collected from the HBV infected subjects or prepared by site-directed mutagenesis of a molecular clone, were cloned into the pCR-Script Amp cloning vector (Stratagene, La Jolla, CA, USA) and used for assessing the analytical performance of the HepB Typer-Entecavir kit. International standard for HBV DNA (PHD801, SeraCare, Gaithersburg, MD, USA) was purchased from SeraCare for calibrating the concentration of the reference standards. Mutations were introduced into the codons rt184, rt202 and rt250 using the Altered Sites II in vitro mutagenesis system (Promega, Madison, Wi, USA). Representative test results of the HepB Typer-Entecavir kit and direct sequencing were presented in supplementary Figure 1.
Substances for the interference test
To examine the effects of different interfering substances that might be present during use of the HepB Typer-Entecavir kit on its performance, test DNA samples in HBV negative human serum were spiked with anticoagulants (wafarin, heparin, EDTA, or sodium citrate), antiplatelet agent (aspirin, ticlopidine), anticoagulants-preservatives (acid citrate dextrose (ACD), citrate phosphate dextrose (CPD), or citrate phosphate dextrose with adenine (CPDA)), antiviral compounds (adefovir, lamivudine, or entecavir), or DNA extraction agent (ethanol; test concentration is 5% and 10%, respectively).
Viruses for cross-reaction evaluation
To evaluate potential cross-reaction, human papillomavirus (HPV), herpes simplex virus (HSV) 1, HSV-2, and human T-lymphotropic virus (HTLV) were obtained from American Type Culture Collection (ATCC). Hepatitis C virus (HCV), hepatitis A virus (HAV), adenovirus, and varicella zoster virus (VZV) were obtained from National Institute for Biological Standards and Control (NIBSC). Human immunodeficiency virus (HIV) was obtained from SeraCare. Viral DNA was determined quantitatively by a branched DNA (bDNA) assay (Versant™ 3.0; Bayer Healthcare LLC Diagnostic Division, Tarrytown, NY, USA), which has a detection limit of 2,000 copies/mL. To test the effect of other viral sequences on performance of HepB Typer-Entecavir kit, some test DNA samples in HBV negative human serum were spiked with HPV, HSV-1, HSV-2, HTLV, HCV, HAV, Adenovirus, VZV, or HIV (50,000 copies/mL, respectively).
Precision evaluation
The test was conducted by 20-day consecutive analysis of each 24 replicates with two concentration HBV samples (1,500 and 50,000 copies/mL) for determination of intra-day and inter-day precision. The 24-replicate comprises 2 independent runs more than 4 hours apart of two-replicates performed by 3 operators in two laboratories (Genematrix, Inc. Yongin lab. and Genematrix, Inc. Seongnam lab.).
RFMP and direct sequencing analyses for testing HBV ETV resistance mutations
Genotypic analysis of HBV ETV-resistant mutations (rt184, rt202, and rt250) was performed using the HepB Typer-Entecavir kit according to the manufacturer's recommendations, (GeneMatrix, Inc., Korea) similar as previously described.22,23 As summarized in Supplementary Table 1, the released diagnostic fragments from enzymatic digestion consist of unique sizes for codons rt184, rt202, and rt250 leading to easy identification of sequence variation in multiple codons in a single mass spectrum. The genotypic analysis by HepB Typer-Entecavir kit was compared with direct sequencing analysis. Direct sequence analysis was performed by ABI PRISM 310 Genetic Analyzer (Applied Biosystems, New York, NY, USA) and HBV genotype was assigned by web-based NCBI retroviruses genotyping analysis of the obtained sequences (http://www.ncbi.nlm.nih.gov/projects/genotyping). All tested samples were genotype C.
Statistical analysis
A limit of detection test was performed by probit analysis to compare sensitivity between the RFMP and direct sequencing assays using the statistical package SPSS version 13 (SPSS Inc., Chicago, IL, USA).
RESULTS
Assessing the lower limit of detection
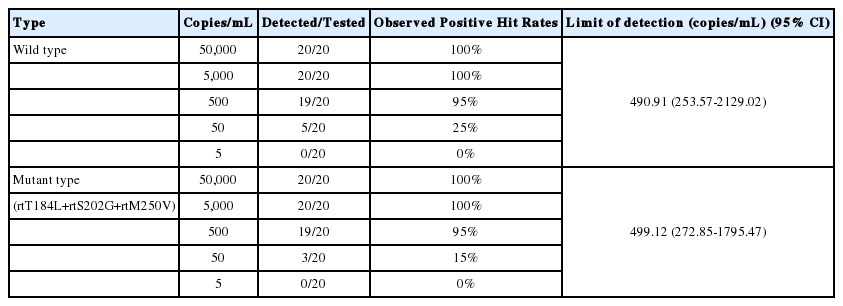
HBV negative serum samples spiked with HBV reference standard from a molecular clone of the wild type and ETV resistance mutant HBV to yield final concentrations of 5, 50, 500, 5,000, and 50,000 HBV DNA copies/ml were used to assess the lower limit of detection of the HepB Typer-Entecavir kit. Analysis of the dilution series of wild and mutant type HBV reference standard revealed detection limits of 490.91 and 499.12 copies per ml, respectively by Probit analysis (Table 1).
Effect of interfering substances
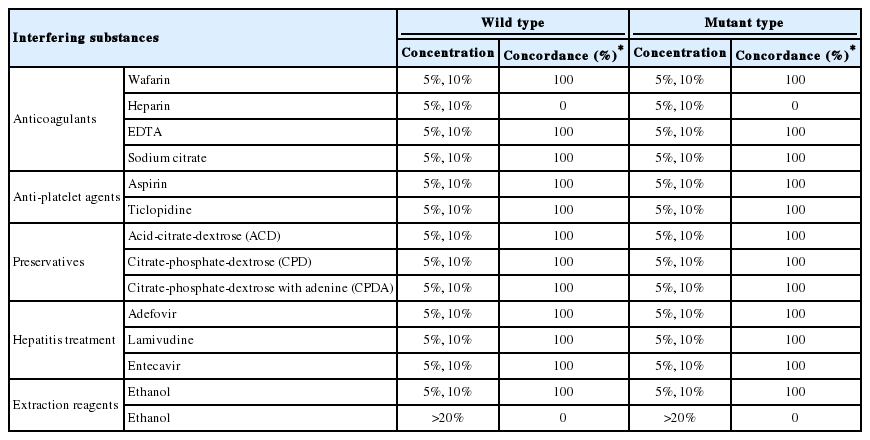
To determine the effect of different interfering substances on assay performance, serum samples containing HBV reference standards at a titer of 1,500 copies/mL were spiked with a wafarin, heparin, EDTA, sodium citrate, aspirin, ticlopidine, ACD, CPD, CPDA, adefovir, lamivudine, entecavir or ethanol as described in Materials and Methods. For each sample, HepB Typer-Entecavir kit data from samples spiked with the various interfering substances were compared to the non-spiked samples. Each biochemical was tested in triplicate. None of the HBV-negative samples spiked with a potentially interfering substance gave a false-positive test result. No negative impact of elevated levels of various interfering substances was observed except the heparinized samples and 20% ethanol spiked samples (Table 2).
Cross-reaction evaluation
To determine the effect of clinically relevant viruses on assay performance, serum samples with or without HBV reference standard (50,000 copies/mL) were spiked with HPV, HSV-1, HSV-2, HTLV, HCV, HAV, adenovirus, VZV, or HIV as described in Materials and Methods. Each pathogen was tested in triplicate. None of the HBV-negative samples spiked with clinically relevant viruses gave a false positive test result. None of clinically relevant viruses affected the HBV-positive samples, irrespective of wild and mutant genotypes (Table 3).
Precision evaluation
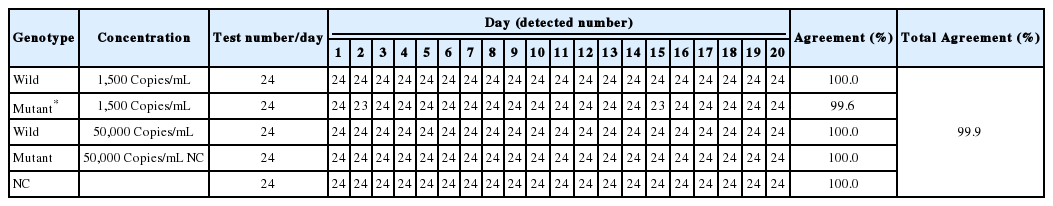
The precision test was conducted by 20-day consecutive analysis at two concentration HBV samples (1,500 and 50,000 copies/mL) of each 24 replicates comprising 2 independent runs of two-replicates performed by 3 operators in 2 laboratories. Precision values were calculated from precision (%) = [(number of true positive + number of true negative)/total sample number] ×100. As shown in Table 4, the total precision conducted by the four reference standards was 99.9%, showing that the HepB Typer-Entecavir kit method has highly reliable reproducibility and repeatability (Table 4).
Comparison of RFMP and direct sequencing assays for HBV ETV resistance mutations
All 396 clinical samples from 108 patients were analyzed by the HepB Typer-Entecavir kit and direct sequencing assays. Codons rt184, rt202 and rt250 were analyzed for the presence of ETV resistance mutations. In the 170 HBV negative specimens (HBV DNA <60 IU/mL), both assays showed HBV DNA negativity in all samples tested. In the 226 HBV DNA positive specimens, the HepB Typer-Entecavir kit detected mutant virus with codons rt184, rt202 and rt250 in 0.4% (1/226), 0.4% (1/226) and 0.9% (2/226) of the sera, respectively. In a total of 396 tested serum specimens, the overall concordance rates were 99.5% between the HepB Typer-Entecavir kit and direct sequencing assays. Concordance rates were 99.7% (395/396) at codon rt184, 99.7% (395/396) at codon rt202, and 100% (396/396) at codon rt250 (Table 5), respectively. Both assays showed identical base substitution and amino acid composition in these positions. Compatible rates were observed 0.3% (1/396) at codons rt184 and rt202, respectively. Two compatible cases, codons rt184 and rt202 mutant harboring patients, had rtM204I/V mutations with HBV DNA level of 1,190 and 7,720 copies/mL, respectively. In this case, HepB Typer-Entecavir kit detected more mixture samples than direct sequencing assay. This sample (mixed-type) at codon rt184 containing double mutations in single codon was identified only by HepB Typer-Entecavir kit, as a result of the inability of the direct sequencing assay to determine the only wild type in the clinical samples. As shown in Figure 1, rt184T/L mixtures detected by HepB Typer-Entecavir kit could be scored as only rt184T (ACT) by direct sequencing. Of these, the HepB Typer-Entecavir kit showed a superior ability for detecting minor virus populations compared with direct sequencing. Overall, HepB Typer-Entecavir kit had 99.5% agreement with direct sequencing when only the detection of mutant virus was considered (Table 5).

Comparison of results obtained using the HepB Typer-Entecavir kit and direct sequencing assays in 396 clinical specimens
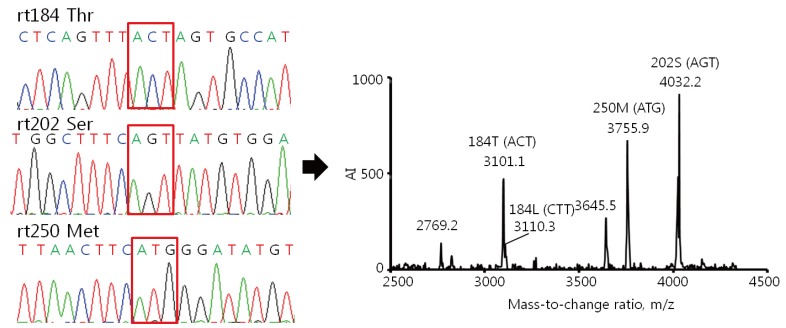
Comparison of the results of the HepB Typer-Entecavir kit and direct sequencing assays for detecting mixed genotypes. Sera were taken from patients infected with HBV and carrying entecavir resistance mutations, and examined using the HepB Typer-Entecavir kit and direct sequencing assays. (A) Molecular masses of 2769.2/3101.0 and 3046.7 represent Thr (ACT) and Leu (CTT) of codon rt184, respectively. However, the direct sequencing assay determined only the Thr (ACT) of codon rt184 in the clinical samples. Molecular mass of 4032.2 represents Ser (AGT) of codon rt202. Molecular masses of 3645.5/3755.9 represent Met (ATG) of codon rt250. Each codon is indicated by a red box in the sequencing chromatogram. AI, absolute intensity; m/z, mass-to-charge ratio.
DISCUSSION
Multi-drug resistance to LMV and ADV is becoming prevalent due to sequential treatment of LMV followed by ADV. ETV displays antiviral activity against both LMV-resistant and ADV-resistant HBV.19,24,25 A previous study showed that 79% of LMV-resistant patients had undetectable HBV DNA levels by bDNA assay at 24 weeks of ETV therapy and that HBV DNA was undetectable by PCR assay in 26% of patients at 48 weeks.11 These results suggest a low response to ETV in patients with LMV resistance and in those with a limited response to ADV. As a consequence, ETV-resistance mutations analysis may be a major tool both for clinicians to monitor treatment and for researchers to consequences for disease outcome.
HepB Typer-Entecavir kit exploits differences in the molecular masses of oligonucleotides comprising the ETV resistance variations in rt region. RFMP assay, which used in HepB Typer-Entecavir kit, is based on mass spectrometric analysis of small DNA fragments, including sites of mutation.22,23,26-33 Mass spectrometry assesses directly the mass of the PCR product, whereas other technologies only measure indirectly PCR products, either through hybridization or by sequencing reactions, which use PCR products as templates. Procedures have been developed in which PCR products are used as templates to which oligonucleotide primers are hybridized, base-extended and then analyzed by mass spectrometry.34 These assays can be useful, but they fail to employ one of the key advantages of mass spectrometry that the analysis of PCR products can be direct. In HepB Typer-Entecavir kit, mass of PCR products is determined directly, rather than their identity being interpreted on the basis of fluorescent or radioactive reporter tags. Both DNA strands can be analyzed in parallel, providing a level of internal confirmation not achievable by other methods. The use of a type IIS restriction enzyme makes the assay independent of restriction sites within the HBV genome and suitable for many different viral genotypes because these enzymes cleave DNA at a fixed distance from the recognition sites incorporated into the amplification primers.
The study reported here assessed the performance of the HepB Typer-Entecavir kit under a variety of analytical conditions. The detection limit of the HepB Typer-Entecavir kit was found to be as low as 500 copies/mL. Presence of anticoagulants, antiplatelet agent, anticoagulants-preservatives, antiretroviral compounds, or ethanol had no significant effect on the accuracy of the kit. As expected, samples anticoagulated with heparin gave negative results, most likely due to interference of heparin with the amplification of HBV DNA.35,36 No significant difference in results was observed in the present study between samples anticoagulated with any anticoagulants except heparin. Nevertheless, careful attention should be given to prompt specimen processing after blood collection in order to optimize assay results. Similarly, presence of other viruses including HPV, HSV-1, HSV-2, HTLV, HCV, HAV, Adenovirus, VZV, and HIV did not cross-reaction with the assay. The cross-reactivity test from samples spiked into clinically relevant viruses suggests that HepB Typer-Entecavir kit is eligible to co-infected patients. The precision test conducted by repetitive analysis of a total of 2,400 replicates with various concentrations of reference standards showed 99.9% agreement, demonstrating that the HepB Typer-Entecavir kit provide highly precise and reproducible genotype results for HBV entecavir resistance mutation.
The performance of the HepB Typer-Entecavir kit for detecting mutations related to ETV drug-resistances was compared with direct sequencing for 396 clinical samples from 108 HBV infected patients. The HepB Typer-Entecavir kit successfully identified genotypic changes tested in clinical samples. The prevalence of ETV resistant mutations in our study was found to be 4.6% (5/108), similar to those reported in previous studies.37,38 Compared with direct sequencing, the HepB Typer-Entecavir kit exhibited 99.5% concordance (Table 5). Specially, the HepB Typer-Entecavir kit outperformed direct sequencing for the detection of single and double mutations in compatible samples. The HepB Typer-Entecavir kit detected more mutant viruses with codons rt184 (1/396) and rt202 (1/396) of the sera, suggesting high sensitivity of the HepB Typer-Entecavir kit in terms of detecting minority variants. Hence, the discrepancy among the two assays may be due to a lower sensitivity of minor mutant of direct sequencing. However, a further study would be required to validate the improved ability to detect mutant virus of HepB Typer-Entecavir kit with more closely monitored samples from increased subjects.
In the previous study, the ability of RFMP method to detect minor virus populations with the LMV, ADV and ETV resistant mutations was superior to that of TRUGENE sequencing.39 All discordance between the two assays related to the detection of additional viral mutation by HepB Typer-Entecavir kit, which was missed by direct sequencing. The minor populations shown in the RFMP were further confirmed by the clonal sequencing, the RFMP assay was in excellent agreement with clonal sequencing (data not shown). These results might have been caused by the different analytical sensitivities of two assays. Viral breakthrough could not predict based only on the presence of the mutant virus and the results of this highly sensitive molecular method should be interpreted carefully, as antiviral drugs could be effective despite the presence of mutants.39 A 5-fold predominance of the drug resistant mutants over wild-type virus was significantly associated with viral breakthrough.30 Therefore, the lack of ability to detect both the wild-type and mutant virus populations could be disadvantage. Direct sequencing generally cannot detect a minor virus population that comprises <20% of the total population,40 whereas RFMP can detect a minor population that comprises only 1% of the total population.28 The previous study has reported that RFMP was superior to PCR-RFLP for distinguishing mixed populations in six of 40 patients22 and in another study that RFMP indicated the presence of additional mutant virus populations compared to INNO-LiPA HBV DR in four of 60 patients.28
In summary, the HepB Typer-Entecavir kit which has recently acquired KFDA approval for in vitro diagnostic use showed the robust analytical performance as well as capability of reliably and reproducibly detecting the entecavir resistance mutations including codons rt184, rt202 and rt250 in clinical samples. Therefore, this assay would be clinically useful in diagnosis of entecavir resistance-associated mutations in chronic hepatitis B.
Acknowledgements
This work was supported by a grant under the Bilateral International Collaborative R&D Program from the Ministry of Trade, Industry and Energy, Republic of Korea.
Notes
The authors have no conflicts to disclose.
Abbreviations
ADV
adefovir
CHB
Chronic hepatitis B
ETV
entecavir
HBV
Hepatitis B virus
HCC
Hepatocellular carcinoma
LMV
lamivudine
MALDI-TOF
matrix-assisted laser desorption ionization time-of-flight
RFMP
restriction fragment mass polymorphism
References
SUPPLEMENTARY MATERIALS
Supplementary Table 1
Expected masses of oligonucleotides resulting from restriction enzyme digestion of PCR products
Supplementary material Table 1 can be found via http://e-cmh.org/suppl/chm-19-4-399-s01
Supplementary Figure 1
Representative results of the HepB Typer-Entecavir kit and direct sequencing assays. The MALDI-TOF MS spectra results and direct sequencing results from the wild type and mutant reference standard are shown. All mutations identified by direct sequencing were concordant with those obtained using the HepB Typer-Entecavir kit. AI, absolute intensity; m/z, mass-to-charge ratio.
Supplementary material Figure 1 can be found via http://e-cmh.org/suppl/chm-19-4-399-s02