Radiomics and radiogenomics of primary liver cancers
Article information
Abstract
Concurrent advancements in imaging and genomic biomarkers have created opportunities to identify non-invasive imaging surrogates of molecular phenotypes. In order to develop such imaging surrogates radiomics and radiogenomics/imaging genomics will be necessary; there has been consistent progress in these fields for primary liver cancers. In this article we evaluate the current status of the field specifically with regards to hepatocellular carcinoma and intrahepatic cholangiocarcinoma, highlighting some of the up and coming results that were presented at the annual Radiological Society of North America Conference in 2017. There are an increasing number of studies in this area with a bias towards quantitative feature measurement, which is expected to benefit reproducibility of the findings and portends well for the future development of biomarkers for diagnosis, prognosis, and treatment response assessment. We review some of the advancements and look forward to some of the exciting future applications that are anticipated as the field develops.
Introduction
Clinical imaging modalities, such as computed tomography (CT), positron emission tomography, or magnetic resonance (MR) imaging, are critical cornerstones in diagnosis and management of modern oncology [1-3]. The maturity of these modalities and protocols of implementation have reached such a degree of sophistication, that properly obtained studies can supplant the need for a tissue biopsy in some diseases, such as the Liver Imaging Reporting And Data System for hepatocellular carcinoma (HCC) [4,5]. Even though the diagnostic protocol for liver cancer is increasingly refined, the prognosis of liver cancers remains unsatisfactory, which is the second most common cause of death from cancer worldwide [6]. Moreover, it is highlighted that variable biologic behavior of liver cancers affects patients’ survival, and the tumoral heterogeneity can be also an important clue in determining the likelihood of a clinical response to treatment [7,8]. Thus, the trend of diagnosis and treatment for liver cancers is changing to be determined by significant biological and genomic characteristics of the tumors, and imaging can be also a method to find these characteristics [9-12].
In contrast to serologic markers, the majority of imaging findings reported from radiologic studies involves qualitative or descriptive features. Moreover, the interobserver variability when radiologists define the imaging findings remains an unresolved issue [13]. Recently, new research methods to explore imaging biomarkers through objective and quantitative analyses of medical images including radiologic studies, have been developed into an academic discipline that studies the association of the imaging parameters with patient information, and several studies have been published in a variety of carcinomas [14,15].
However, this state-of-the-art method is still in its infancy, especially in the liver cancer research; various attempts have been made and presented in the annual meeting of Radiological Society of North America (RSNA) in 2017. This review article focuses on the preliminary clinical application of radiomics, and radiogenomics (or imaging genomics) for primary liver cancers, such as HCC and intrahepatic cholangiocarcinoma (ICC).
What are radiomics, radiogenomics, and imaging genomics?
Radiomics is a method of research that extracts quantitative radiologic data from medical images (radiomic data) and explores the correlation with clinical outcomes [14]. Radiogenomics (or imaging genomics) also aims to identify relationships between semantic as well as quantitative image data with genome and molecular measurements in order to construct association maps that can in turn be correlated with outcome or other clinical measures [15-17]. A requisite for both radiomics and radiogenomics is the identification of an objective manner to transform the imaging features into digital data, which is also important for insuring reproducibility. The general radiomics workflow involves, 1) acquisition of images, 2) identification of region of interest (by human or machine learning), 3) segmentation of region of interest (by automatic, semiautomatic, or manual drawing), 4) extraction of imaging features, and 5) mining the data to develop model to predict clinical outcomes (Fig. 1).
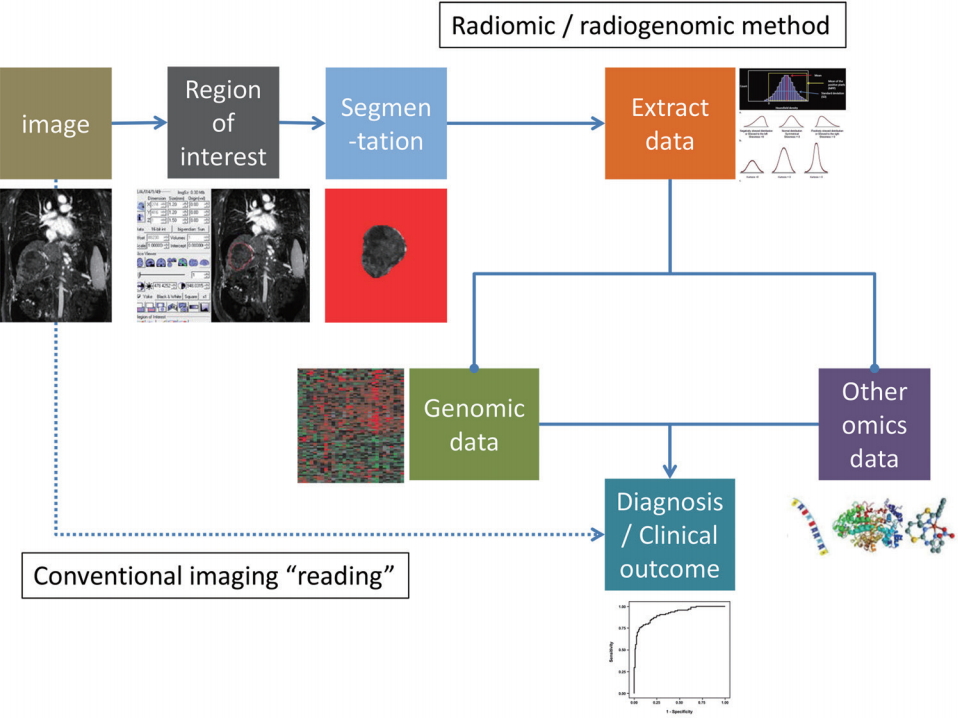
The process of radiomics and radiogenomics. In comparison to the current conventional imaging study interpretation, radiomic and radiogenomic approaches require multiple processing steps (automated as well as semi-automated steps including registration, segmentation, region of interest selection, measurement, etc.). As the fields develop and methodologies become more standardized these steps may also become “implicit” in the image processing component, similar to the processing of raw computed tomography (CT) or magnetic resonance (MR) data before transmission to clinical PACS. Selected icons adapted and reprinted from Lubner et al.[19], with permission from Radiological Society of North America (RSNA). PACS, picture archiving and communication system.
For radiomics and radiogenomics, it is possible to analyze the medical images archived in the imaging server of the hospital retrospectively, and the more imaging database has sufficient data, the more accurate predictive radiomic model can be made. However, the variability of imaging quality and technical parameters for image acquisition may influence the image features and results of predictive models.
Radiological imaging traits: quantitative versus qualitative methods
The gold standard of radiologic image interpretation continues to be expert (radiologist) based reads, in spite of digital acquisition (or conversion) of clinical imaging data. In order to convert these images into parametric data objectively, it should be helped by specific analytic method, feature analysis (FA). FA is a quantitative method of objectively assessing tumor heterogeneity or parenchymal change in specific disease by exploring the distribution and connection of pixel or voxel gray levels in the CT image [18]. There are several parts, such as the gray-level frequency distribution from the pixel intensity histogram in a given region of interest, a co-occurrence matrix which considers the location of pixels and spatial relationship between pixels, and neighborhood gray-tone difference matrices which examine location and relationship between three or more pixels. In addition, model-based and transform-based methods are used for FA (Fig. 2) [19].
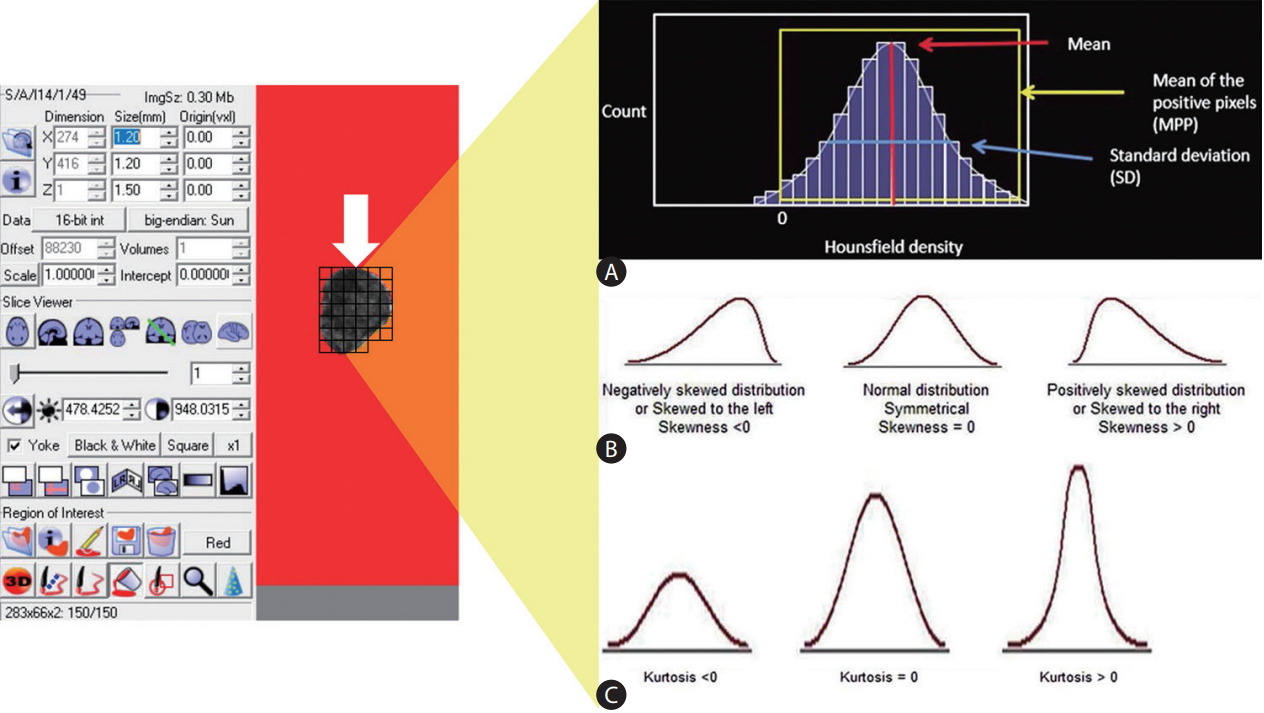
An example of acquiring texture parameters through feature analysis. Following selection of an appropriate the region of interest (white arrow) of an image, (A) a histogram of gray-level intensity and distribution can be drawn. From the histogram, several statistical measures including mean, mean of the positive pixel, standard deviation can be calculated (A). The distributions of the measurements can further be characterized in terms of their (B) skewness, and (C) kurtosis. Modified and reprinted from Lubner et al.[19], with permission from Radiological Society of North America (RSNA).
Performance of quantitative analyses has multiple prerequisites. First of all, reasonable segmentation of the tumor should precede the FA. Segmentation is the most critical component because the subsequent feature data are generated from the segmented volumes, but it is still challenging due to indistinct border of the tumors [16]. At this step, it is vulnerable to be disturbed by reviewer’s subjectivity; therefore, automatic or semi-automatic segmentation methods are preferred to manual methods [20], especially for liver cancers of which tumor-parenchyma differentiation may be not clear compared with lung and breast cancers. Second, denoising and gray-level standardization can affect the analytic result, especially second- and high-order statistics, thus these should be performed properly in the pre-processing steps to aid in reproducibility of the analysis.
One advantage of descriptive or qualitative features is that they frequently describe physiological or biological processes. For example, there is a largely accepted explanation for arterial phase enhancement of HCC masses in contrast-enhanced CT and MR images. The development of HCC frequently involves neovascularization of unpaired arteries that do not have associated portal tracts [21,22], thus increasing arterial flow through the unpaired artery as compensation for decrease of portal flow in the tumor. Consequently, contrast media goes into the tumor early on the arterial phase and goes out of the tumor through veins on following phases, called as wash-out [23]. Nowadays, these findings have been considered as hallmarks for imaging diagnosis of HCC [4,5]. While it is often reassuring when such features have a demonstrated (patho-)physiological description, a challenges is the difficulty with inter-observer , and even intra-observer, variability [5].
Biologic and genomic characteristics of liver cancers
HCC is the most common primary cancer in the liver. It is known to be a quiet killer with minimal symptoms at the early stages of disease, frequently occurring in the context of hepatitis infections and cirrhosis leading to end-stage liver disease, in addition to largely unknown tumor genetic factors. Current staging systems, such as Barcelona Clinic Liver Cancer staging system, do not take into account the histologic and molecular characteristics of the tumor [9].
According to recent studies, HCCs exhibit intra-individual and inter-individual genetic heterogeneity. Predicting biologic behavior of HCC is essential for an appropriate treatment, and investigators have found that patient prognosis is not only related to size of tumor and multiplicity, but also the pathologic features such as histologic grade of tumor, microvessel density, microscopic vascular invasion (MVI), and epithelial-mesenchymal transition (EMT) [24-26].
Histologic grade of HCC accounts for a combination of structural and cellular features and is a significant predictor. According to a meta-analysis of 114 related articles, poor tumor differentiation correlated with worse prognosis [27]. However, there was a limitation of divergences and inaccuracies led by a non-standardized grade distribution. Microvessel density, which allows semi-quantitative assessment of tumor neovascularization (CD31, CD34, and von Willebrand Factor), is known as a significant prognostic factor. Vascular endothelial growth factors (VEGFs) and hypoxia-inducible factors (HIFs) are also negative prognostic markers related to reduce overall survival and recurrence-free survival after tumor resection [28].
MVI is another well-known factor suggesting poor prognosis. In the large-scale retrospective study which observed transplanted patients’ survival, the authors revealed a doubling in the hazard of death was associated with the presence of MVI [29]. EMT is another pathologic feature to predict prognosis and it increases invasiveness of tumor and metastatic potential [30,31].
In terms of genomic markers, in order to accurately predict therapeutic response of targeted therapy based on tumor molecular profiles, there have been efforts to identify critical molecular subclasses with different prognostic consequence, and associated genetic or epigenetic drivers of specific subclasses.
Some investigators have proposed molecular classifications of HCC using mRNA-based gene expression based on biological processes (e.g. proliferation) and protein markers (e.g. EpCAM and cytokeratin 19 [CK19]) [32]. Villanueva et al. focused on both the tumor and nontumoral-adjacent cirrhotic tissue in order to identify a genomic signature predictive of patients’ outcome, and they suggested a model for prediction of recurrent HCC combining clinical, pathology, and genomic data [33].
ICC is another aggressive primary liver cancer and new studies are identifying pathologic biomarkers that reflect the biological behavior of the tumor. Overexpression of epidermal growth factor receptor (EGFR) was an independent prognostic parameter along with tumor stage [34]. EGFR, VEGF, and human epidermal growth factor receptor 2 were also significantly related to macroscopic type, nodal metastasis, lymphatic vessel invasion, perineural invasion and tumor stage [35].
KRAS mutation was found in up to 40% of cholangiocarcinoma, but predominantly in perihilar and distal tumors [36]. Because direct therapeutic inhibition of KRAS is difficult, targeted therapy aims at modulation of downstream signaling along the KRAS pathway. Recently, Graham et al. identified FGFR2 translocation in ICC, and it could be benefited from FGFR-targeted therapy [37].
Like HCC, microvessel density and EMT may play a role as biomarkers to indicate prognosis of ICC. Microvessel density and lymphatic microvessel density were frequently displayed in advanced ICC, and the patients of which microvessel density was high had an inferior curative resection rate and frequently developed recurrence [38]. In addition, the high expression of N-cadherin and S100A4, the pathologic evidences of EMT, were independently significant prognostic factors in ICC [39].
Where do we stand on the radiomics and radiogenomics for primary liver cancer?
There are limited quantitative analyses of imaging studies in the hepatic tumors to date although some researchers have focused on texture analysis. Most of the studies have focused on the relationship between imaging traits and clinical characteristics including survival, recurrence, and treatment response after chemotherapy. In contrast, some studies dealt with the relationship between the genomic signatures and imaging findings (without texture analysis); therefore, radiogenomic analysis for liver cancer is very rare so far (Table 1) [40,41].
The majority of the studies have centered on HCC, and been based on CT texture with only some studies which investigated the role of MRI [16]. At the 2017 RSNA Conference, there were several presentations about radiomics of HCC using MRI, especially gadoxetic acid-enhanced MRI.
HCC
The first radiogenomic study of HCC was performed in 2007 [10] and involved measurement of quantitative as well as semantic imaging features. They found that combination of 28 imaging traits could reconstruct the variation of all 116 gene models, about 80% of global gene expression profiles, revealing cell proliferation, liver synthetic function, and patient prognosis. Among the imaging traits, the presence of internal arteries and the absence of a hypodense halo were associated with MVI genomic signatures that were comprised of genes involved in cell proliferation (CDK, CDC20, MCM5) and matrix invasion (ADAMTS1, MMP14, SPARCL1). On a subsequent study, the CT-based imaging traits were evaluated to predict the presence of MVI on pathologic specimen, and additional imaging trait, the absence of tumor-liver difference, was considered the third imaging trait for “radiogenomic venous invasion” imaging signature as well as the established two imaging traits. This signature was not only accurate to diagnose MVI but associated with lower overall survival [11].
Villanueva et al. [33] investigated the genomic features of HCC and peritumoral tissue that were associated with patients’ outcome, who tried to explore the relationship between imaging trait and genomic signatures [42]. Patients who underwent pre-operative CT or MR imaging before surgery as well as transcriptome profiling were assessed 11 qualitative and four quantitative (size, enhancement ratios, wash-out ratio, tumor-to-liver contrast ratio) imaging traits. Several imaging traits including infiltrative pattern and macrovascular invasion were associated with gene signatures of aggressive HCC phenotype such as proliferative signatures and CK19 signature.
More recently a radiomics signature derived from texture analysis was proposed to be a surrogate for MVI in HCC, prediction of patients’ survival, and early recurrence [43-45]. This study involved 28 HCC patients who underwent resection and evaluated some imaging textures and “delta features” defined as the absolute difference or the ratio calculated from all pairs of imaging phases (e.g. precontrast minus arterial phase Hounsfield units) [43]. Among 464 features, single and delta features between arterial and portal-venous phases were robust to detect MVI. Another research group published that contrast-enhanced MR-based texture analysis could differentiate pathologic grade [46]. Forty-six patients who performed contrast-enhanced MRI before tumor resection were enrolled and arterial phase images were used for analysis. Histological low-grade HCCs had increased mean intensity and decreased gray-level run-length non-uniformity in four directions. This suggest that internal heterogeneity could be an imaging feature reflecting high-grade HCC, which has poor clinical prognosis. Recently, a study group tried to explore the CT-based radiomics data which related to key genomic information such as doxorubicin-resistant genes (TP53, TOP2A, CTNNB1, CDKN2A and AKT1), and they identified radiomics signatures to successfully discriminate these chemo-resistant genes [41].
The majority of studies to date have focused on CT imaging features, however, now several MR imaging based studies are emerging and features associated with MVI have been found, such as decreased apparent diffusion coefficient (ADC) on diffusion weighted imaging [47], peritumoral enhancement, non-smooth tumor margin [48], and peritumoral hypointensity on hepatobiliary phase imaging following gadoxetic acid injection [49]. There are some MR imaging sequences, such as diffusion, that can provide information that is not measurable by CT [50], thus radiomic MR features are of great interest. However, MR imaging is more vulnerable to imaging artifacts such as motion and magnetic susceptibility, thus reproducibility between different scanners and even the same scanner is a non-trivial challenge. Additionally the signal intensity of MR imaging is not easy to be digitalized compared to CT imaging (Hounsfield units), which is easy to be modulated and standardized for radiomic analyses.
Last year at the RSNA Conference (2017), three results on MR-based radiomics analysis for HCC were presented. Hui et al. sought to predict early recurrence after resection using MR imaging [51]. The study involved 57 patients that were separated into cohorts with early recurrence or not, using texture analysis of contrast and entropy in the arterial phase with 79% accuracy. Chen et al. showed texture analysis on the perfusion MR imaging could be associated with Edmudson-Steiner grades, and skewness of area under curve map was revealed as a potential marker to predict histologic grades [52]. Hectors et al. analyzed MR imaging texture on multiparametric MRI including blood oxygenation level dependent imaging and dynamic contrast enhanced-MRI, and investigated the correlation with HIF-1α and gene expression of Wnt target GLUL, pharmacological target FGFR4, stemness markers EpCAM and KRT19 and immune checkpoint PDCD1 [53].
Intrahepatic cholangiocarcinoma
To date there has only been one radiogenomics ICC study. 25 enrolled patients with histologically proven ICC were evaluated according to CT-based textures and immunochemistry findings suggesting tissue hypoxia, such as EGFR, VEGF, CD24, TP53, MDM2, MRP-1, HIF-1α, CA-IX, and GLUT1 [12]. Following multiple variable regression, three imaging phenotypes, entropy, correlation, and homogeneity, were significantly associated with EGFR expression and correlation phenotype was associated with VEGF expression level although visual (qualitative) features were not associated with all of hypoxia markers. Another retrospective study tried to reveal which CT imaging phenotypes were associated with patient’s survival and tumor genetics. CT imaging features were semantic and not quantitative; for example, central fibrosis, well-defined border, homogeneity of enhancement, central necrosis, liver surface retraction, vascular encasement, tumor thrombus, satellite lesions, intrahepatic metastasis and lymphadenopathy [54]. They found that necrosis, satellite nodule, and vascular encasement might be associated with a higher risk of death. However, they could not find the relationship between imaging features and genetic mutation such as IDH1 and KRAS gene.
Several published studies investigated imaging findings as related to patient outcome after resection. Asayama et al. evaluated delayed phase CT exams in a cohort of 32 patients and observed that the degree of enhancement correlated with the amount of fibrous stroma and the frequency of perineural invasion [55]. The survival of patients was also associated with the degree of enhancement [55]. Lee et al. explored that diffusion-weighted image could predict patients’ survival after resection [56]. Multivariate regression test revealed that degree of diffusion restriction was associated with patients’ survival, as well as histologic differentiation and intrahepatic metastasis on pathology.
A quantitative MR imaging study by Pandey et al. [57], explored imaging feature changes in 72 patients with unresectable ICC and underwent two MRI examinations before and after chemotherapy. Paradoxically, ADC values on the baseline study were lower and percent of viable tumor volume was higher in the group with longer survival (>10 months); it was hypothesized that radiologically viable tumor was more responsive to the chemotherapy.
Future perspectives of radiomics: strengths and weaknesses
The preceding studies suggest that radiomic and radiogenomic approaches can be useful in gaining deeper insights into tumor genetics, biology, and clinical outcome. MR in particular likely has many new applications with the development of functional pulse sequences in addition the anatomical information, although standardization of protocols and image scans will continue to be a challenge for quantitative imaging analysis. It is conceivable that in the future, we will be able to use radiomics-based decision making tool to diagnose incidental hepatic lesion or to decide which treatment is best to the disease. Artificial intelligence and deep learning will also likely have roles to play in the future of radiomics and radiogenomics, particularly with regards to registration and segmentation of imaging studies.
Quantitative imaging approaches, including imaging texture analysis, reflect a departure from the historical, largely subjective approach for medical image interpretatin. The merit of this new method is that it is able to get digital information (data) from medical imaging and when performed under appropriate protocols, is more robust and reproducible than strictly human based interpretations, i.e., radiologist. A potential advantage is that information that may be “hidden” can be discerned with quantitative approaches. On the same token, there is a knowledge base that the radiologist has that radiomic based approaches have not yet achieved. Although there are too many variables to reliably predictive the future, it is likely that at a minimum these automated (or semi-automated) approaches will support the diagnostic and decision-making processes in medical imaging.
Nevertheless, there are remaining issues for clinical use. First, reproducibility is a very important issue [14]. This is related to many factors, such as imaging acquisition protocol, method of segmentation, method for extracting imaging features, and acquisition of clinical and genomic data. With respect to tumor markers, the standard reporting recommendations (REporting recommendations for tumour MARKer prognostic studies, REMARK) has been established for over 10 years [58]. In a similar vein, standardization of analysis methodologies in radiomics and radiogenomics will be necessary if these methods are to gain broad adoption in the imaging community. Second, estimating appropriate sample size is another important issue. While larger data sets and “big data” provide great promise to identify new predictors of survival and outcome, associations do not imply causality [17]. Third, although many radiomic studies have been implemented retrospectively, ideally a prospective study design is preferable. Additionally multisite hospitals may help increase the study size number and statistical power but at the cost of increased variability in standardization of protocols and data processing [17].
Conclusion
While oncologic radiomics and radiogenomics are in early stages of development there is much to be gained from continued investigations in order to maximize the utility of non-invasive diagnostic tests and potential molecular and genomic correlates of imaging features. Towards this end they may also advance the objectives of precision medicine as well as more cost effective healthcare.
Notes
Authors’ contribution
WKJ and NJ outlined the manuscript, content, with subsequent revisions and edits.
WKJ drafted the manuscript.
All authors edited the manuscript.
All authors agreed on content.
Notes
Conflicts of Interest: The authors have no conflicts to disclose.
Abbreviations
ADC
apparent diffusion coefficient
CK
cytokeratin
CT
computed tomography
EGFR
epidermal growth factor receptor
EMT
epithelialmesenchymal transition
HCC
hepatocellular carcinoma
ICC
intrahepatic cholangiocarcinoma
MR
magnetic resonance
MVI
microscopic vascular invasion
RSNA
Radiological Society of North America
VEGF
vascular endothelial growth factor

