No association between the IL28B SNP and response to peginterferon plus ribavirin combination treatment in Korean chronic hepatitis C patients
Article information
Abstract
Background/Aims
There are few available data regarding the association between the single nucleotide polymorphisms (SNPs) of the gene encoding interleukin 28B (IL28B) and a sustained virologic response (SVR) to peginterferon (PEG-IFN) plus ribavirin (RBV) therapy in Korean chronic hepatitis C patients.
Methods
This was a retrospective cohort study of 156 patients with chronic hepatitis C virus (HCV) infection who received combination treatment of PEG-IFN plus RBV. Blood samples from these patients were analyzed to identify the IL28B SNPs at rs12979860, rs12980275, rs8099917, and rs8103142. Association analyses were performed to evaluate the relationships between each IL28B SNP and SVRs.
Results
Seventy six patients with HCV genotype 1 and 80 with genotype non-1 were enrolled. The frequencies of rs12979860 CC and CT genotypes were 90.4% and 9.6%, respectively; those of rs12980275 AA and AG genotypes were 87.2% and 12.8%, respectively; those of rs8099917 TT and TG genotypes were 92.3% and 7.7%, respectively; and those of rs8103142 TT and CT genotypes were 90.4% and 9.6%, respectively. Among the patients with HCV genotype 1, the SVR rates were 69.7% and 80.0% for rs12979860 CC and CT, respectively (P=0.71). Among the HCV genotype non-1 patients, SVR rates were 88.0% and 100% for rs12979860 CC and CT (P=1.00), respectively.
Conclusions
Genotypes of the IL28B SNP that are known to be favorable were present in most of the Korean patients with chronic hepatitis C in this study. Moreover, the IL28B SNP did not influence the SVR rate in either the HCV genotype 1 or non-1 patients. Therefore, IL28B SNP analysis might be not useful for the initial assessment, prediction of treatment outcomes, or treatment decision-making of Korean chronic hepatitis C patients.
INTRODUCTION
Hepatitis C virus (HCV) is a major cause of chronic viral hepatitis behind the hepatitis B virus. Although a small proportion of the patients infected with HCV are spontaneously cleared, 50-90% of patients are destined to suffer from chronic infection, which is associated with variable degrees of hepatic inflammation and fibrosis progression. Ten to 40% of patients with chronic HCV infection develop cirrhosis, and the estimated incidence of hepatocellular carcinoma is 1-5% per year among cirrhosis patients.1 Interferon-based therapy is the current standard of care for chronic HCV infection and has been proven to reduce the risks of cirrhosis and hepatocellular carcinoma.2,3,4,5,6 With peginterferon (PEG-IFN) and ribavirin (RBV) combination therapy, the sustained virologic response (SVR) rate is approximately 40-50% for HCV genotype 1 patients and approximately 70-80% HCV genotype non-1 patients in Western countries.7,8,9 In contrast, the SVR rates are higher among Korean patients and reaching approximately 70% in genotype 1 and 80-90% in genotype non-1 patients, respectively.10 Recently, genome-wide association studies reported that the interleukin (IL) 28B single nucleotide polymorphism (SNP) is associated with the SVR to the PEG-IFN and RBV combination. Therefore, determination of the IL28B SNPs may be useful for predicting treatment responses and managing the patients with chronic HCV infection.11,12 However, in Asia, the majority of patients possess the genotypes that are favorable for interferon-based therapy, including the representative SNPs rs12979860 and rs8099917, which may limit the usefulness of the IL28B SNP in treatment response prediction.13,14 Meanwhile, a study of Caucasian patients reported that rs8099917 SNP among the group of patients with the non-favorable SNP rs12979860 is helpful in the prediction of SVR.15 Therefore, we intended to evaluate the potential additional benefits of several promising IL28B SNP analyses in Korean patients with chronic HCV infection.
PATIENTS AND METHODS
Study subjects
This is a retrospective cohort study that included all patients with chronic HCV infection who were treated with the PEG-IFN alfa-2a or -2b plus RBV combinations as initial antiviral treatments at Asan Medical Center from July 1, 2004 to June 30, 2011. The inclusion criteria were 18 to 75 years old, compensated liver status, positivity for both anti-HCV and HCV RNA, and a treatment duration ≥80% of the planned treatment duration (48 weeks for HCV genotype 1, and 24 weeks for HCV genotypes 2, 3, and 6). The exclusion criteria were other previous antiviral treatments for HCV infection, co-infection with HBV or HIV, and hepatocellular carcinoma or liver transplantation. Another study cohort included patients with spontaneous HCV clearance, which was defined by positivity for anti-HCV and negativity for HCV RNA without previous antiviral treatment, from the Asan Medical Center between Jan 1, 2000 and June 30, 2011. All study patients were of Korean ethnicity. This study was approved by the Institutional Review Board at Asan Medical Center.
Treatment protocol
For the patients with genotype 1, PEG-IFN α-2a (Pegasys®; Roche, Basel, Switzerland) 180 µg/week, or PEG-IFN α-2b (PegIntron®: Schering Plough, Kenilworth, NJ, USA) 1.5 µg/kg/week was injected, and RBV (Robavin®; Shinpoong, Seoul, Korea) 1,000 mg (for body weights <75 kg) or 1,200 mg (for body weights ≥75 kg) was administered for 48 weeks. The patients with genotype 2, 3, or 6 were treated with PEG-IFN α-2a or α-2b at the same doses described above and RBV at 800 mg for 24 weeks. HCV RNA was quantified (Roche AMPLICOR HCV Test v2.0; Roche, Mannheim, Germany) prior to treatment and at weeks 12, 24 and 48 for both genotypes and at week 72 for genotype 1. The selection of the PEG-IFN α-2a or α-2b treatment was at the discretion of clinical physician. Doses were modified according to the contemporary treatment guidelines when side effects developed.
Clinical data collection
The patients' clinical data were collected from electronic medical records. A sustained virologic response (SVR) was defined as a documented serum level of HCV RNA that was undetectable 24 weeks after the cessation of treatment.
Single nucleotide polymorphism genotyping
Serum and buffy coat samples were collected and stored at -80℃ in a refrigerator bank. Genomic DNA was extracted from the peripheral blood buffy coats using the QIAamp DNA Blood Maxi Kit (QIAGEN, Valencia, CA, USA) according to the manufacturer's instructions. Four target SNPs (rs12979860, rs12980275, rs8099917, and rs8103142) were amplified using primer sets. Polymerase chain reactions (PCRs) were performed using a PTC-200 thermal cycler (MJ Research, Waltham, MA, USA). Cycle sequencing was performed on an ABI Prism 3130XL Genetic Analyzer using the BigDye Terminator Cycle Sequencing Reaction Kit (Applied Biosystems, Life Technologies, Carlsbad, CA, USA).
Statistical analysis
To compare the baseline characteristics of the antiviral treatment responder and non-responder groups, chi-square tests or Fisher's exact tests were used for categorical variables, and Student's t-tests or Mann-Whitney U-tests were used for continuous variables as appropriate. Chi-square tests or Fisher's exact tests were used in the association analyses of relationships of the SVR and each SNP genotype. All tests were two-sided, and P values below 0.05 were considered statistically significant. The linkage disequilibrium structure was analyzed with Haploview 4.2 (Broad Institute, Cambridge, MA, USA). Univariate and multivariate binary logistic regression analyses were used to identify the factors that were predictive of SVR.
RESULTS
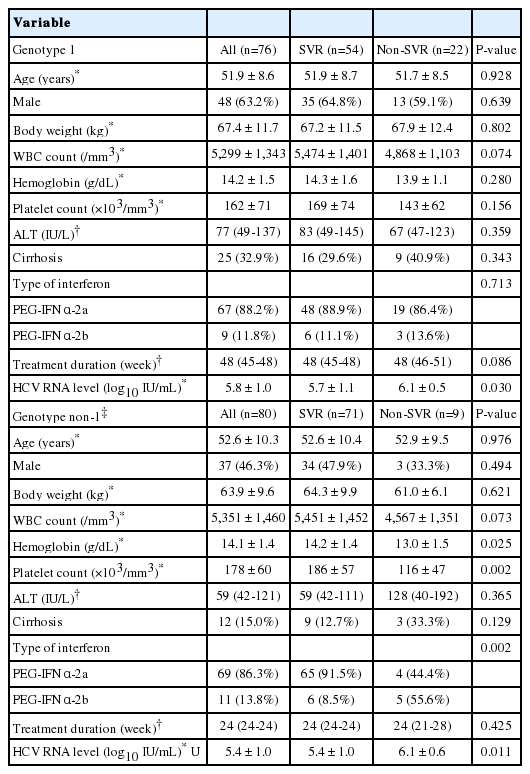
Baseline characteristics of the patients
Of the 239 chronic hepatitis C patients who met the eligibility criteria, 156 patients agreed to enroll in the study and to provide blood samples following antiviral treatment. Seventy-six and 80 patients had the HCV genotypes 1 and non-1, respectively. The baseline characteristics, including age, gender, body weight, white blood cell count, hemoglobin, platelet count, ALT level, cirrhosis, type of interferon, and treatment duration, were similar between the genotype 1 SVR and non-SVR groups. The initial HCV RNA level was higher among the non-SVR group than the SVR group. Among the genotype non-1 patients, the initial HCV RNA level and the proportion of peginterferon α-2b treatments were higher in the non-SVR group than in the SVR group (Table 1). A total of 27 patients were enrolled in the spontaneous HCV clearance cohort.
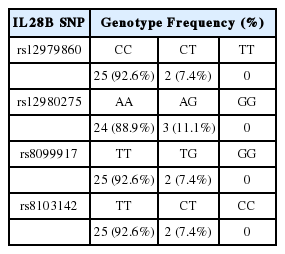
Genotype distribution according to IL28B SNP
In the treatment cohort, the majority of the patients exhibited favorable genotypes at each SNP. The frequencies of rs12979860 CC and CT were 90.4%, and 9.6%, respectively. The frequencies of rs12980275 AA and AG were 87.2% and 12.8%, respectively; the frequencies of rs8099917 TT and TG were 92.3% and 7.7%, respectively; and the frequencies of rs8103142 TT and CT were 90.4% and 9.6%, respectively. No patient was homogenous for the unfavorable genotype at each SNP (Table 2). Similarly, in the spontaneous clearance cohort, the majority had favorable genotypes at each SNP (Table 3).

Percentage genotype distributions of the IL28B SNPs rs12979860, rs12980275, rs8099917, and rs8103142 among patients who were treated with PEG-IFN and RBV
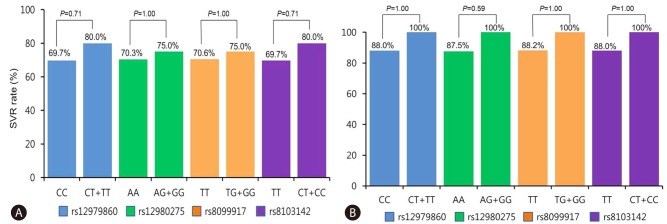
Association between IL28B genotype and SVR
Within the treatment cohort of 156 patients, 125 (80.1%) exhibited SVRs. Among the 76 HCV genotype 1 patients, 54 (71.1%) patients exhibited SVRs. Among the 80 HCV genotype non-1 patients, 71 (88.8%) patients exhibited SVRs. In the HCV genotype 1 group, the SVR rates were 69.7% and 80.0% for the rs12979860 CC and CT groups, respectively; 70.3% and 75.0% for the rs12980275 AA and AG groups, respectively; 70.6% and 75.0% for the rs8099917 TT and TG groups, respectively; and 69.7% and 80.0% for the rs8103142 TT and CT groups, respectively (Fig. 1A). Among the HCV genotype non-1 patients, the SVR rates were 88.0% and 100% for the rs12979860 CC and CT groups, respectively; 87.5% and 100% for the rs12980275 AA and AG groups, respectively; 88.2% and 100% for the rs8099917 TT and TG groups, respectively; and 88.0% and 100% for the rs8103142 TT and CT groups, respectively (Fig. 1B). The SVR rates were not statistically different between the favorable and unfavorable IL28B SNP genotype groups irrespective of HCV genotype.
IL28B haplotype analysis
The linkage disequilibriums of rs12979860 with rs8099917 (r2=0.79), rs12980275 (r2=0.73), and rs8103142 (r2=1.00) were high. Thus, the allele frequencies were nearly identical for the SNPs rs12979860, rs8099917, rs12980275, and rs8103142. Consequently, no effects of including these variants in the haplotype or combination analyses could be expected (Fig. 2).
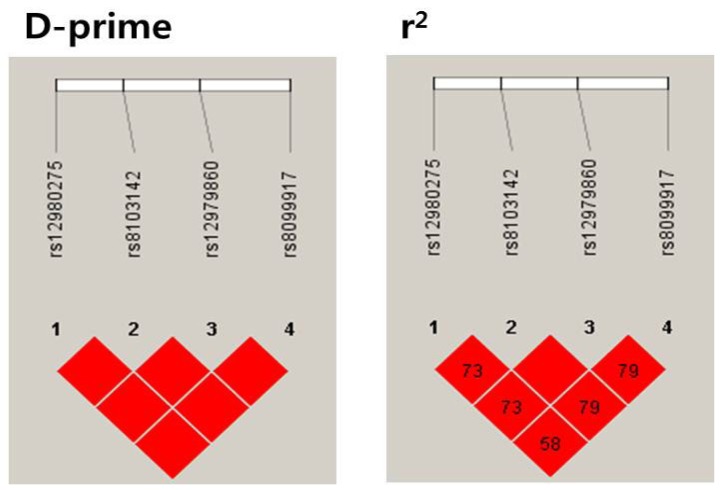
IL28B SNP linkage disequilibria (LDs). D-prime=proportion of the possible LD that was present between the SNPs. D' varies from 0 (complete equilibrium) to 1 (complete disequilibrium). r2=correlations between the SNPs. When r2=1, two SNPs are in perfect LD, and the allele frequencies are identical for each SNP. The values in the box are percentages. Empty spaces in the box indicate values of 100%.
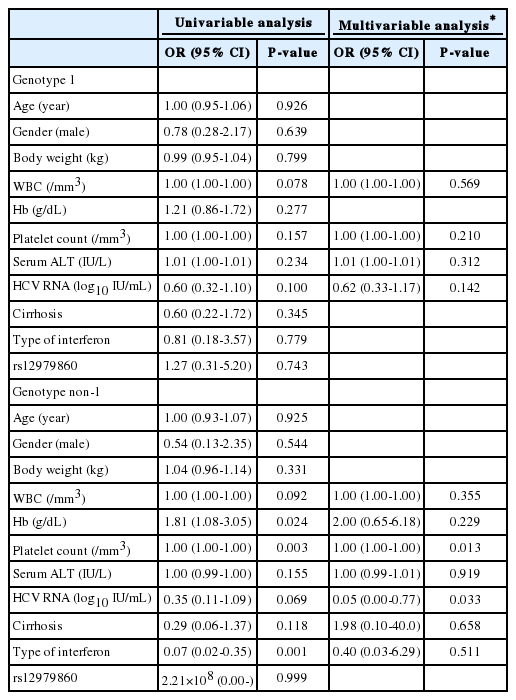
Predictive factor analysis for SVR
Age, gender, body weight, WBC count, hemoglobin, platelet count, serum ALT level, initial HCV RNA level, cirrhosis, type of interferon, and rs12979860 were included in a logistic regression to predict the SVR. Among the HCV genotype 1 patients, no variables were found to be significantly predictive factors in the Univariable analyses. In the multivariable analysis that included the variables for which the P-values were below 0.25 (i.e., WBC count, platelet count, serum ALT level, and initial HCV RNA level), no variables were found to be significant predictive factors for SVR. Among the HCV genotype non-1 patients, platelet count and type of interferon were significant predictors of SVR in the univariable analyses (the P-values were 0.003 and 0.001, respectively). However, the mutlivariable analysis that included the variables for which the P-values were below 0.25 (i.e., WBC count, hemoglobin, platelet count, serum ALT level, initial HCV RNA level, cirrhosis, and type of interferon), no variables were found to significantly predict SVR (Table 4).
DISCUSSION
The present study was performed to address the issue of whether the IL28B SNP influences treatment responses to PEG-IFN plus RBV combination therapy among Korean chronic hepatitis C patients. We evaluated SVRs according to IL28B SNPs that included rs12979860, rs8099917, rs12980275, and rs8103142; all of these SNPs are known in the literature to have treatment-favorable homozygous and treatment-unfavorable hetero-/homozygous genotypes.11,12,16,17,18 All four of the SNPs were strongly linked, and the majority of patients had the favorable homozygous genotypes at each SNP. A small proportion of the patients had unfavorable heterozygous genotypes, and no patient carried unfavorable homozygous genotypes. SVRs were not significantly different between the favorable homozygous and unfavorable heterozygous genotypes. Moreover, the predictive factor analysis did not indicate that the IL28B SNP was associated with SVR.
Because IFN-based therapy can cause several significant adverse effects, the prediction of SVRs to antiviral treatments at the level of individual patients can influence on treatment decisions. Several viral and host factors, such as HCV genotype, pretreatment HCV RNA level, age, gender, and cirrhosis, are known to be predictive of SVR. Additionally, recent genome-wide association studies have suggested that the IL28B SNP is associated with the SVR to IFN-based therapies among chronic hepatitis C patients.
Recent studies have suggested that the SNPs rs12979860, rs8099917, rs12980275, and rs8103142 are correlated with treatment outcome and spontaneous viral clearance.11,12,16,17,18,19,20 The most frequently observed variants, rs12979860 CC and rs8099917 TT, are significantly associated with SVRs to PEG-IFN plus RBV therapy among HCV genotype 1 patients.11,12 Although African Americans exhibit lower SVR rates, this trend persists across all ethnicities, including European Americans, African Americans, and Hispanics. Because the frequency of the rs12979860 CC type is highest among East Asians, the favorable treatment outcomes of East Asians are thought to be related to the high frequency of treatment-favorable SNPs.11 This explanation has been supported by studies of Koreans in which most of the Korean patients had the rs12979860 CC and rs8099917 TT types, and these patients achieved higher SVR rates than did the rs12979860 CT/TT type and rs8099917 TG/GG type patients.13,14,21 The present study also found that the treatment-favorable genotypes were present in approximately 90% of Korean patients. However, our results revealed that the SVR rate of the unfavorable heterozygous genotype group was similar to that of the favorable homozygous genotype group. This finding may be the result of type 2 error due to low statistical power, but it is also possible that an ethnic factor other than the IL28B SNP influences treatment outcomes among Korean patients.
Fischer et al15 reported that rs12979860 and rs8099917 exhibit high linkage disequilibrium and that, in cases of the unfavorable rs12979860 CT/TT type, rs8099917 SNP determination influences the prediction of SVR among HCV genotype 1 patients. However, in the present study, only two of 10 HCV genotype 1 patients with the unfavorable rs12979860 CT/TT type had the discordantly favorable rs8099917 TT type, and all ten of them achieved SVR. Therefore, we were unable to evaluate the additional effects of the analysis of rs8099917 following rs12979860.
IL28A, IL28B, and IL29 encodings recently identified members of the IL-10 cytokine superfamily, type III interferon or IFN-γ. Although previous studies have shown that favorable genotypes of the IL28B SNPs are associated with SVR irrespective of ethnicity, the negative predictive values, i.e., the SVRs associated with the unfavorable homozygous genotype, are still 26-31% among HCV genotype 1 patients. This finding indicates that a significant proportion of patients achieve SVR despite having the unfavorable alleles.22 Treatment adherence is one of important host factor that is predictive of SVRs to IFN-based therapy.23 We included patients in the treatment cohort only if their treatment durations were ≥80% of the planned treatment duration and did not evaluate dose reduction rates among the patients. Thus, it is possible that the patients with unfavorable IL28B SNP genotypes could achieve a high SVR rate if treatment adherence exceeds 80% not only in terms of treatment duration but also in terms of PEG-IFN and RBV doses. Notably, the SVR rates of HCV genotype 1 Korean and Taiwanese patients in clinical trials have been reported to be as high as 80%, but the SVR rates in clinical practice in Korea vary between 35~74%.10 Given that there is a gap in treatment adherence between clinical trial and practice, the efforts enhancing strict treatment adherence might have overcome the differences due to IL28B SNPs.
In the spontaneous HCV clearance cohort, the proportion of patients with the favorable rs12979830 CC type was 93%, which was similar to that of the treatment cohort. The high proportion of favorable IL28B SNP genotypes may have influenced the spontaneous HCV clearance, but it is also possible that another host factor influenced the clearance. Futures studies examining the effects of IL28B SNPs on spontaneous HCV clearance in acute hepatitis C Korean patients are required.
The present study has several limitations. First, not all eligible patients were enrolled for the collection of blood samples for the analyses of the IL28B SNP; thus, it is possible that sampling bias was present. However, the patient number of this study is the highest among relevant studies that have been published in Korea. Therefore, the possibility of sampling error in this study might be lower than that of other studies. Second, because approximately 90% of the patients had favorable IL28B SNP genotype, a larger sample size is needed to discriminate the effects of the less common unfavorable genotypes on SVR. The small sample size of this study might have resulted in type 2 error and a false conclusion that SVRs were not different between the groups of patients with different IL28B SNPs.
In conclusion, the known favorable genotypes of the IL28B SNP were present in the majority of the Korean chronic hepatitis C patients. However, the IL28B SNP did not influence the SVR rates of either the HCV genotype 1 or non-1 patients. Therefore, IL28B SNP analysis might be not useful for the initial assessment of Korean chronic hepatitis C patients, the prediction of the treatment outcome, or treatment decision making.
Acknowledgments
The authors thank Kyung-Hwa Hong for help with the data collection.
Notes
The authors have no conflicts to disclose.
Abbreviations
ALT
alanine aminotransferase
IL28B
interleukin 28B
Hb
hemoglobin
HCV
hepatitis C virus
PEG-IFN
peginterferon
RBV
ribavirin
RNA
ribonucleic acid
SNP
single nucleotide polymorphism
SVR
sustained virologic response
WBC
white blood cell