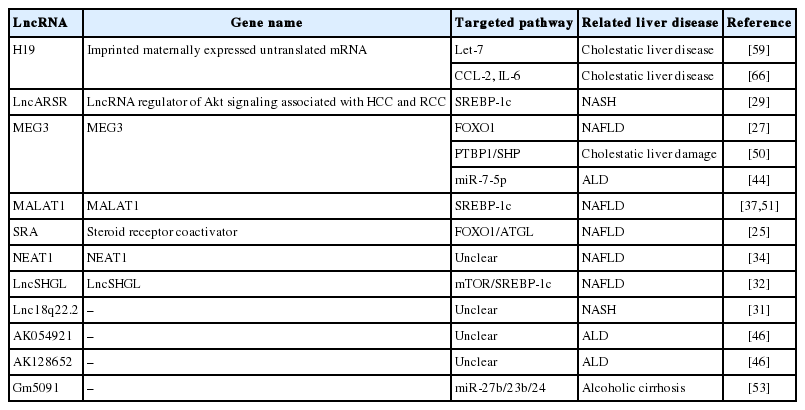
Long non-coding RNAs in liver diseases: Focusing on nonalcoholic fatty liver disease, alcohol-related liver disease, and cholestatic liver disease
Article information
Abstract
Long non-coding RNAs (lncRNAs), a class of transcribed RNA molecules with the lengths exceeding 200 nucleotides, are not translated into protein. They can modulate protein-coding genes by controlling transcriptional and posttranscriptional processes. The dysregulation of lncRNAs has been related to various pathological disorders. In this review, we summarized the current knowledge of lncRNAs and their implications in the pathogenesis of three common liver diseases: nonalcoholic fatty liver disease, alcohol-related liver disease, and cholestatic liver disease. Future studies to further define the role of lncRNAs and their mechanisms in various types of liver diseases should be explored. An improved understanding from these studies will provide us a useful perspective leading to mechanism-based intervention by targeting specific lncRNAs for the treatment of liver diseases.
INTRODUCTION
Overview of long non-coding RNA (lncRNA)
The central dogma “DNA-RNA-protein” indicates that key genetic information is stored in protein-coding genes; however, only a small percentage of the human transcriptome is translated into proteins, leaving the majority of the transcripts undefined. In recent decades, studies have focused on the remaining genome and found that up to 90% are non-coding RNAs (ncRNAs) [1,2]. These ncRNAs were initially considered as junk RNA or transcriptional noise; however, lately, they are involved in various cellular and disease processes, with emerging evidence of their interactions with each other as a complex regulatory network [3]. In general, ncRNAs are divided into two groups depending on the length of the nucleotides, small ncRNAs (<200 nucleotides), and lncRNAs (>200 nucleotides) [4].
Classifications of lncRNAs
LncRNAs are transcribed by RNA polymerase II or III, mostly are 5’-capped, 3’-end poly-adenylated, and multi-exonic [5,6]. They exhibit poorer conservation than protein-coding genes [7]. Most lncRNAs are folded after the transcription to form the tertiary structure, which impacts their functions [8,9]. Based on their genomic location, lncRNAs are classified into the following categories; 1) intergenic lncRNA, located in between two protein-coding genes, transcribed in the same direction; 2) intronic lncRNA, located in the intronic regions and transcribed entirely from introns of a protein-coding gene; 3) sense lncRNA, transcribed from the same strand and the same direction as the surrounding proteincoding genes; they can be overlapping with exonic or intronic regions or cover the entire protein-coding sequence through an intron; 4) antisense lncRNA, transcribed from the opposite strand of surrounding protein-coding genes, also can be both exonic and intronic; and 5) bidirectional lncRNA, located within 1 kb of the promoter region of a protein-coding gene, but transcribed from the opposite strand [10,11]. There are also some other criteria to classify lncRNAs based on their transcript properties, location in known genomic annotations, regulatory elements, and functions [12].
Functions of lncRNAs
LncRNAs are involved in several biological processes such as the shaping of chromatin, replication, transcription, splicing, translation, and post-translational modification of proteins [13,14]. For example, signal lncRNAs are only expressed at a specific time and localized to certain subcellular structures where they exert their functions [15]. Decoy lncRNAs binds to regulatory factors such as transcription factor, RNA-binding protein, and chromatin modifier, thereafter altering their original biological activity [15]. Guide lncRNAs regulate gene activation or repression by forming ribonucleoprotein complexes and mediating their localization to specific targets [15]. Scaffold lncRNAs are similar to guide lncRNAs, but they affect the molecular component of the complex itself [16,17]. Enhancer lncRNAs are produced from enhancer elements and influence the activation of target genes [18]. Moreover, as a reservoir, some lncRNAs are processed to generate microRNAs (miRNAs) [19,20]. Some lncRNAs can bind miRNAs and function as a sponge for certain miRNAs to block their activity, which involves regulating the protein translation of the target mRNA [21,22]. The main functions of the lncRNAs are shown in Figure 1.
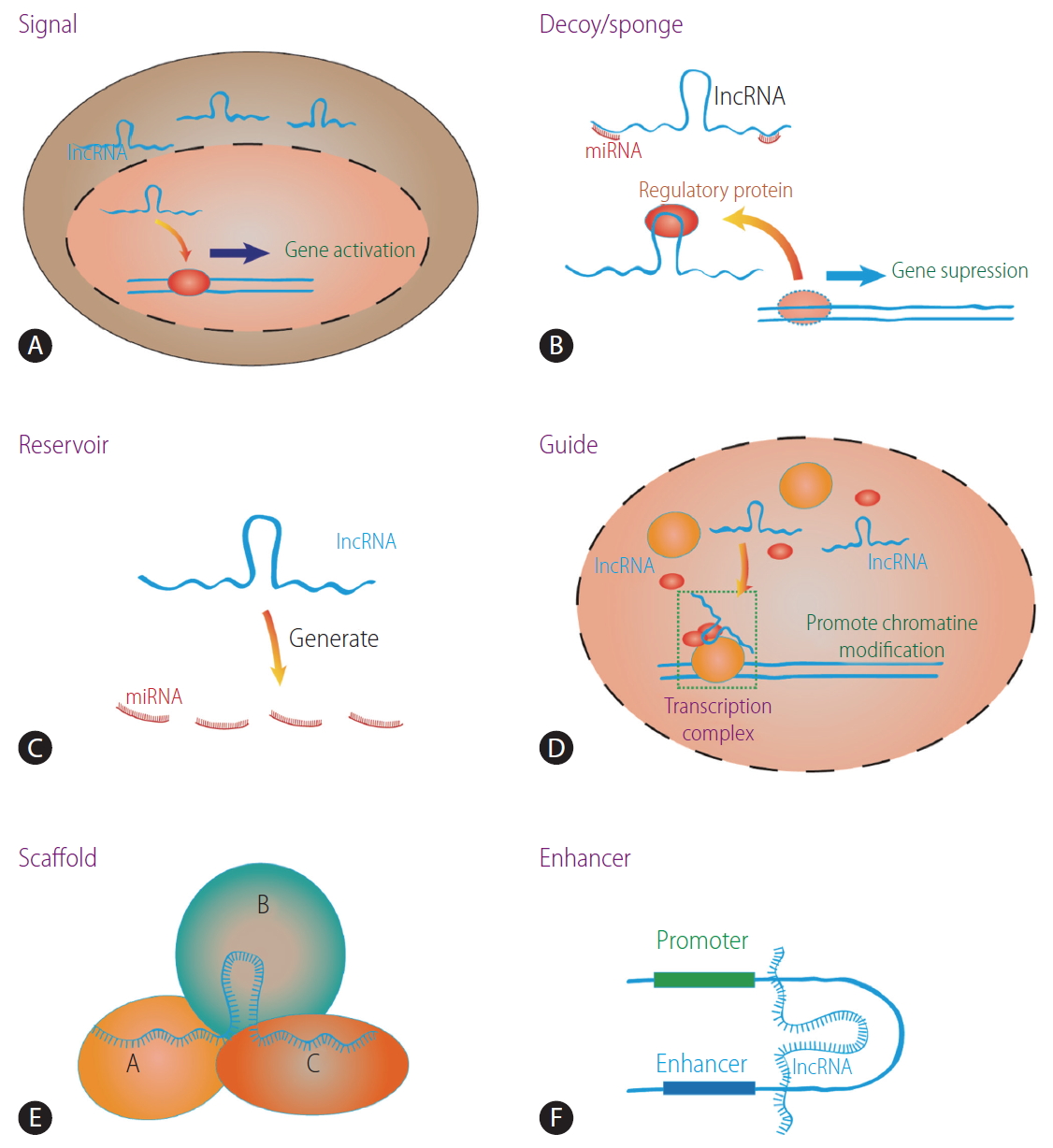
The main biological functions of lncRNA. (A) Signal lncRNA promotes gene expression. (B) Decoy lncRNA binds to regulatory factors and affects gene expression. LncRNA can sponge miRNA and affects its function. (C) Reservoir lncRNA generates miRNA. (D) Guide lncRNA takes ribonucleoprotein complexes to specific target. (E) Scaffold lncRNA combines other molecules to form complex. (F) Enhancer lncRNA increases target gene expression. lncRNA, long non-coding RNA; miRNA, microRNA.
Because lncRNAs are involved in a wide variety of important cellular processes, dysregulation of lncRNAs has been related to various pathological disorders. In this review, we summarized the current knowledge of lncRNAs and their implications in the pathogenesis of 3 common liver diseases, nonalcoholic fatty liver disease (NAFLD), alcohol-related liver disease (ALD), and cholestatic liver disease.
A CRITICAL ROLE OF LncRNAs IN NAFLD
NAFLD comprises a spectrum of histopathological disorders ranging from simple steatosis, steatohepatitis, advanced fibrosis, and cirrhosis [23]. Emerging studies have shown that lncRNAs are implicated in the pathogenesis of NAFLD through their roles in lipid and glucose metabolism.
LncRNA steroid receptor RNA activator (SRA)
SRA is involved in glucose and lipid metabolism [24,25]. A study showed that the expression of liver adipose triglyceride lipase (ATGL) is induced in SRA knockout mice, and liver SRA and ATGL expression are inversely regulated under fasting conditions [25]. Forkhead box protein O1 (FOXO1) is a key transcription factor in the regulation of gluconeogenesis and the insulin response in the liver, which plays an important role in glucose homeostasis [26]. FOXO1 mainly targets insulin signaling and regulates metabolic homeostasis in response to oxidative stress, which also promotes the transcription of ATGL. By inhibiting the ability of FOXO1, lncRNA SRA inhibits ATGL promoter activity and regulates ATGL expression;a potential mechanism underlies NAFLD pathogenesis.
LncRNA maternally expressed gene 3 (MEG3)
LncRNA MEG3 is involved in hepatic glucose metabolism [27]. MEG3 is upregulated in mice fed with a high-fat diet, while knockdown of MEG3 remarkably abolishes hepatic triglyceride (TG) accumulation, up-regulates glycogen content, and promotes glucose tolerance [27]. Palmitate, oleate or linoleate also increases MEG3 expression level in primary hepatocytes. Histone deacetylases (HDACs) are a class of enzymes involved in expression of DNA, which catalyze the removal of acetyl groups from lysine residues in both histone and non-histone proteins. Knocking down of HDAC3 using siRNA or its inhibitor (RGFP966) significantly promotes MEG3 expression level, suggesting the role of histone acetylation in regulating its expression [27]. Upregulation of MEG3 also increases FOXO1 expression and hepatic insulin resistance [28].
LncRNA activated in renal cell carcinoma with sunitinib resistance (lncARSR)
The mRNA levels oflong non-coding RNA activated in renal cell carcinoma with sunitinib resistance (lncARSR) both in the serum and liver are significantly increased in NAFLD patients compared with the healthy controls (HC) [29]. Hepatic lncARSR is also induced in mice fed with methionine choline deficient diet. Overexpression of lncARSR enhances while knockdown of lncARSR ameliorates hepatic lipid accumulation in vivo and in vitro . Transient overexpression of lncARSR using AAV8-lncARSR virus markedly induces the expression of lncARSR and causes significant increase of intracellular TG contents in HepG2 cells [29]. LncARSR overexpression also induces the expression of genes associated with lipogenesis, such as sterol regulatory element-binding protein 1 (SREBP-1c), fatty acid synthase (FASN), and stearoyl-CoA desaturase. Knockdown of SREBP-1c by short hairpin RNA is shown to block the effect of lncARSR on lipogenesis. In addition to SREBP-1c, overexpression of lncARSR also increases the level of phosphorylated Akt, and its effects was inhibited in the presence of LY294002, the phosphatidylinositol 3-kinase (PI3K) inhibitor. These data suggest the role of lncARSR-PI3K-Akt axis in regulating lipogenesis through SREBP-1c [29].
LncARSR also modulates cholesterol metabolism in vitro and in vivo [30]. LncARSR overexpression significantly elevated the serum total cholesterol (TC), low density lipoprotein cholesterol, and hepatic TC. The expression of genes related to cholesterol biosynthesis such as Srebp2, HMG-CoA reductase (Hmgcr), and HMG-CoA synthase also increases, while the expression of Cyp7a1, the rate limiting enzyme of the conversion of cholesterol to bile acid (BA), decreases, suggesting the modulation of hepatic cholesterol biosynthesis by lncARSR [30]. The effect of lncARSR on cholesterol biosynthesis is dependent on Akt/SREBP-2 pathway [30].
Lnc18q22.2
Lnc18q22.2, a liver-specific lncRNA, is markedly induced in the liver tissue of nonalcoholic steatohepatitis (NASH) patients and correlated with NASH severity [31]. The silencing of lnc18q22.2 expression results in reduced growth in HepG2 and immortalized human hepatocyte cells and promotes cell death in Huh7 and Hep3B cells; suggesting its crucial role for growth and viability of hepatocytes [31]. Lnc18q22.2 is directly or indirectly involved in numerous essential biological processes in hepatocytes, including oxidation reduction, translation elongation, and regulation of cell death. Genes negatively regulated by lnc18q22.2 are enriched in the process of oxidation reduction; the finding which is consistent with the observation that NAFLD is often accompanied by increased oxidative stress [31]. One of the most enriched pathways which is affected in the loss of lnc18q22.2 function is the regulation of cell death and apoptosis which may underlie the cell viability. Hepatocyte apoptosis is a major feature in NASH and in fact several anti-apoptosis genes were down-regulated after lnc18q22.2 knockdown, including MCL1, BCL2L1, BCL2L2, BFAR, CARD10, IGFIR, and MKL [31].
LncRNA suppressor of hepatic gluconeogenesis and lipogenesis (lncSHGL)
The expression of lncSHGL in mouse and its human homologous lncRNA B4GALT1-AS1 are decreased in the liver of obese mice and NAFLD patients [32]. Fasting hyperglycemia is improved at the 4th and 7th day after the injection of adenovirus overexpressed lncSHGL in mice fed with high fat diet. Insulin tolerance test and hyperinsulinemic euglycemic clamp shows the improvement of global insulin resistance with the reduction in hepatic gluconeogenesis at the 7th day post injection. LncSHGL overexpression also reduces hepatic TG and serum insulin levels. Consistent with enhanced insulin sensitivity, hepatic lncSHGL overexpression repressed the lipolysis of white adipose tissue and reduced serum free fatty acid levels in high fat diet fed mice [32]. LncSHGL overexpression also increases phosphorylated Akt with reduced protein levels of phosphoenolpyruvate carboxykinase, glucose 6-phosphatase and FASN. Further mechanistic studies reveal that lncSHGL promotes Akt activation and FOXO1 nuclear exclusion in calmodulin-dependent manner. LncSHGL recruits heterogeneous nuclear ribonucleoprotein A1 to enhance calmodulin mRNA translation, activates the PI3K/Akt pathway, and inhibits the mammalian target of rapamycin (mTOR)/SREBP-1c pathway to prevent hepatic steatosis [32].
Nuclear enriched abundant transcript 1 (NEAT1)
LncRNA NEAT1 is involved in many diseases. As an oncogenic lncRNA, NEAT1 is remarkably up-regulated in lung cancer and breast cancer cells and promotes the breast cancer cell proliferation and DNA synthesis [33]. NEAT1 also plays an important role in NAFLD pathogenesis. The levels of NEAT1 are increased in NAFLD in vivo and in vitro [34]. Overexpression of NEAT1 increases both mRNA and protein level of acetyl coA carboxylase and FASN. It also leads to the induction of phosphorylated-mTOR and phosphorylated-p70S6K1. The effect of NEAT1 on acetyl co-A carboxylase (ACC) and FAS expression is ameliorated in the presence of rapamycin, an inhibitor of mTOR/S6K1 pathway [34]. The inhibition of mTOR/S6K1 pathway mimics the effect of NEAT1 knockdown on the expression of ACC and FAS mRNA levels. Knocking down of NEAT1 significantly improves NAFLD phenotypes in the NAFLD rat model; suggesting the important role of the NEAT1/mTOR/S6K1 axis in the pathogenesis of NAFLD [34].
Metastasis-associated lung adenocarcinoma transcript (MALAT1)
MALAT1 plays an important role in diabetic complications and in the pathogenesis of diabetes-related microvascular disease and diabetic retinopathy [35]. MALAT1 stimulates the production of inflammatory cytokine in the endothelial cells in the presence of high glucose [36]. MALAT1 expression is dose-dependently increased in HepG2 cells and primary mouse hepatocytes when they are exposed to different doses of palmitate [37]. The MALAT1 levels are also elevated in livers from ob/ob mice [37]. Knockdown of MALAT1 significantly reduces the palmitate-induced TG and cholesterol accumulation in HepG2 cells and primary mouse hepatocytes. The reduction of MALAT1 expression abolishes palmitate-enhanced nuclear SREBP-1c protein level but had no effect on SREBP-1c precursor and SREBP-1c mRNA in HepG2 cells and mouse hepatocytes. MALAT1 knockdown also results in a significant decrease in the expression of SREBP-1c target genes. MALAT1 upregulates SREBP-1c expression by stabilizing SREBP-1c protein in hepatocytes by preventing ubiquitin-mediated degradation [37]. Lastly, MALAT1 is also involved in NASH fibrosis. MALAT1 regulates C-XC motif chemokine ligand 5 (CXCL5) expression and the expression of both MALAT1 and CXCL5 is altered during the transition of hepatic stellate cells (HSCs) from a quiescent to activated state [38].
LncRNA H19
H19 as well as genes that are involved in hepatic lipogenesis are upregulated in mice fed with high fat diet [39,40]. H19 directly downregulates the expression of miR-130a which in turn inhibits the expression of peroxisome proliferator-activated receptor γ level in the liver [39]. It also promotes steatosis by regulating transcription factor MLX-interacting protein-like (MLXIPL) or carbohydrate-responsive element-binding protein [40]. H19 also serves as a lipid sensor by synergizing with RNA-binding protein, polypyrimidine tract-binding protein 1 (PTBP1), to modulate hepatic metabolic homeostasis [41]. H19 can interact with PTBP1 to increase SREBP1 protein cleavage, stability, and transcriptional activity to activate lipogenesis genes [41].
A CRITICAL ROLE OF LncRNAs IN ALD
ALD occurs in a subset of patients with excessive alcohol intake [42,43]. Similar to NAFLD, ALD also consists of a spectrum of pathological changes ranging from steatosis to fibrosis and cirrhosis. Although the relationship between alcohol consumption and liver disease is well established, severe alcohol-related morbidities develop in only a minority of people who consume alcohol in excess. Inter-individual differences in susceptibility to the toxic effects of alcohol have been extensively studied [43]. Several new lines of evidence demonstrate the important role of lncRNAs in ALD pathogenesis
MEG3
Chronic alcohol consumption induced MEG3 expression in hepatocytes in both in vivo and in vitro models. Knocking down of MEG3 leads to a significant decrease in intracellular lipid accumulation in the presence of ethanol secondary to the inhibition of SREBP-1c and FASN [44]. LncRNAs have been reported to act as decoys to sequester miRNAs to prevent them from binding to targets [45]. There is a negative regulation between MEG3 and let-7c-5p, as MEG3 is a target of let-7c-5p in alcohol-induced liver injury [44]. Overexpression of let-7c-5p Inhibits alcohol-induced steatosis by the regulation of its target gene NOD-like receptor family CARD domain containing 5 [44].
LncRNAs AK128652 and AK054921
Our group has conducted a study with the hypothesis that specific lncRNAs are critical for the progression from excessive drinkers (ED) without underlying liver disease to ALD and their unique role underpins the clinical observation on why only a subset of ED develop ALD. Using the lncRNA microarray global profiling, we identified unique lncRNA signature changes in HC and ED with more differentially expressed lncRNAs in patients with alcoholic cirrhosis [46]. Detailed analysis of these lncRNAs in alcoholic cirrhosis stratified by Child‐Pugh classes demonstrates 244 up‐regulated and 181 down‐regulated lncRNAs compared to HC. The number of uniquely up‐regulated lncRNAs increased from 252 for those in Child-Pugh class A to 1,417 for those in Child-Pugh class C, whereas the number of uniquely down‐regulated lncRNAs increased from 100 in those with Child class A to 1,594 for those in Child class C [46]. We reported 19 unique lncRNAs to be present in the liver on differential expression either >2 or <2 folds change. Among them, lncRNA AK128652 exhibited the highest induction in patients with alcoholic cirrhosis followed by lncRNA AK054921. These two lncRNAs also exhibit prognostic significance on survival in patients with alcoholic cirrhosis; suggesting the unique roles of lncRNAs as potential prognostic biomarkers in patients with ALD [46].
LncRNA Gm5091
Hepatic fibrosis is a result of the dysregulation of the extracellular matrix (ECM) synthesis secondary to HSCs activation. HSCs activated by various toxic stimuli undergo proliferation and myofibroblastic transformation, lose their retinol and produce a considerable amount of ECM, such as α-smooth muscle actin and type I collagen [47,48]. Several lncRNAs have been implicated in the pathogenesis of fibrosis and cirrhosis through their functions as either pro-fibrotic and anti-fibrotic effects [49-52]. LncRNA Gm5091 is enriched in mouse HSC and significantly downregulated in the liver during alcohol-induced hepatic fibrosis [53]. LncRNA Gm5091 negatively regulates cell migration, reactive oxygen species content, interleukin-1β secretion, and expression of collagen I and markers of HSC activation by sponging miR-27b/23b/24 [53].
A CRITICAL ROLE OF LncRNAs IN CHOLESTATIC LIVER DISEASE
Cholestasis is a condition with a decrease in bile flow, either due to impaired secretion by hepatocytes or by obstruction [54]. BAs, synthesized from cholesterol in the liver, play an essential role in eliminating cholesterol and facilitating dietary fat absorption. BA metabolism is strictly controlled by nuclear receptor signaling to coordinate BA synthetic enzymes and transporters [55]. Accumulation of BAs in the hepatocytes can cause cell damage leading to inflammation and fibrosis [55]. The nuclear receptor small heterodimer partner (SHP; NR0B2) is an essential component in the negative feedback regulation of BA synthesis. In response to the intrahepatic accumulation of BA, nuclear receptor farnesoid X receptor (FXR) activates SHP to repress the expression of two regulatory enzymes sterol 12α-hydroxylase and cholesterol 7α-hydroxylase for de novo synthesis of BA [56,57]. The activation of SHP also represses the transactivation of basolateral BA transporter of the hepatocytes such as sodium-taurocholate co-transporting polypeptide (NTCP) to block uptake of BA [58]. Recent studies suggest the role of lncRNAs in cholestasis.
LncRNA H19
In the bile duct ligation (BDL) model, the hepatic let-7 family is markedly induced by H19 [59]. H19 promotes the biogenesis and expression of a cluster of let-7 miRNAs, including let-7a-5p, let-7d-5p, and let-7f-5p, in hepatic cholestasis and functionally suppresses their bioavailability. H19 antagonizes the expression of its binding partner, PTBP1, which associates with pre-let-7d and prelet-7a-1 and inhibits let-7 biogenesis but promotes their bioavailability [59]. H19 facilitates hepatic cholestasis by preventing zinc finger E-box binding homeobox 1 (ZEB1) mediated inhibition of epithelial cell adhesion molecule (EpCAM). ZEB1 is a transcriptional repressor that plays a role in epithelial-to-mesenchymal transition during fibrogenesis [60]. H19 induces EpCAM but inhibits ZEB1 expression in the BDL model and overexpression of ZEB1 or knockdown of EpCAM ameliorates H19-induced cholestasis [61].
In another model of cholestasis using multidrug resistance 2 deficient (Mdr2–/–) mice, the expression of hepatic H19 in 100-daysold wild-type mice was very low with a slight but significant increase in male Mdr2–/– mice. However, there was a markedly increase in its expression by 200-folds in female Mdr2–/– mice with more severe cholestasis [62]. Further mechanistic experiments revealed that taurocholate and 17β-estradiol up-regulate H19 expression and that knocking down H19 ameliorates cholestatic liver injury in female Mdr2–/– mice [62].
H19 is also implicated in biliary atresia [63]. Hepatic H19 expression and the levels of serum H19 are associated with the degree of fibrosis in patients with biliary atresia [63]. Additionally, its expression is correlated with the up-regulation of hepatic sphingosine 1-phosphate receptor 2 and sphingosine kinase 2 which are known to attribute to cholestatic liver injury by enhancing cholangiocyte proliferation and inflammatory responses [63-65].
MEG3
MEG3 can interact with PTBP1 as identified using RNA pulldown with biotin-labeled sense or anti-sense MEG3RNA followed by mass spectrometry. Forced expression of MEG3 in hepatocellular carcinoma cells guided and facilitated PTBP1 binding to SHP coding region, resulting in SHP mRNA decay. Transient overexpression of MEG3RNA in vivo in mouse liver caused rapid SHP mRNA degradation and cholestatic liver injury, which was accompanied by the disruption of BA homeostasis, the elevation of liver enzymes, and dysregulation of BA synthetic enzymes and metabolic genes [50]. Summary of lncRNAs and their roles in liver diseases are shown in Table 1.
PERSPECTIVES AND CONCLUSION
Numerous lncRNAs are implicated in the pathogenesis of NAFLD, ALD, and cholestatic liver diseases. The regulatory network of lncRNAs and the underlying mechanisms leading to liver diseases are complicated. Given a large number of lncRNAs and the extensive studies in this area, new lncRNAs with relevance to liver diseases are likely to be discovered. While the field is advancing, there is a critical need to translate the observations in cell lines or animal models and determine if the functions of these lncRNAs recapitulate the disease pathogenesis in humans. Detection of circulating lncRNAs raises the potential for their use as the biomarkers for risk prediction, diagnostic applicability, or predictors of prognosis in liver diseases. It will be of interest to determine the use of specific lncRNAs that can perhaps supplement and eventually replace invasive liver biopsy for diagnostic and prognostic purposes. Modulation of specific lncRNAs especially in mouse models has a profound impact on the liver disease phenotypes, suggesting the therapeutic potentials of lncRNA-based therapy. Despite the considerable promise, there are several challenges on the safety and the potential off-target effects. An improved understanding and knowledge resulting from the intensive studies of lncRNAs in liver diseases will unquestionably provide useful perspectives and generate new information that will eventually translate to novel clinical applications.
Acknowledgements
ZY is supported by NIH K01AA26385, Indiana University Research Support Fund Grant (IU RSFG), and the Ralph W. and Grace M. Showalter Research Trust and the Indiana Institute for Medical Research; SL is supported in part by R01 DK107682, R01 AA025208, U01 AA026917, UH2/UH3 AA026903, VA Merit Award 1I01CX000361 and Indiana Clinical and Translational Sciences Institute, UL1TR002529, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award, Showalter Scholar Indiana University School of Medicine, and IUSM Dean’s Scholar in Medical Research; PK is supported by the grant from Indiana Institute for Medical Research (IIMR).
Notes
Authors’ contributions
SH: Literature review, SH, TZ, YJ: Drafting the manuscript, PK, NH, SL, and ZY: A critical review of the manuscript, SH and SL: Finalizing the manuscript. All authors have read and approved the manuscript for submission.
Conflicts of Interest
The authors have no conflicts to disclose.
Abbreviations
ACC
acetyl coA carboxylase
ALD
alcohol-relatedliver disease
ATGL
adipose triglyceride lipase
BA
bile acid
BDL
bile duct ligation
CXCL5
C-X-C motif chemokine ligand 5
ECM
extracellular matrix
ED
excessive drinkers
EpCAM
epithelial cell adhesion molecule
FASN
fatty acid synthase
FOXO1
Forkhead box protein O1
FXR
farnesoid X receptor
HC
healthy controls
HDACs
histone deacetylases
hnRNPA1
heterogeneous nuclear ribonucleoprotein A1
HSCs
hepatic stellate cells
lncARSR
long non-coding RNA activated in renal cell carcinoma with sunitinib resistance
lncRNA
long non-coding RNA
lncSHGL
long noncoding RNA suppressor of hepatic gluconeogenesis and lipogenesis
MALAT1
metastasis-associated lung adenocarcinoma transcript
Mdr2–/–
multidrug resistance 2 deficient
MEG3
maternally expressed gene 3
miRNA
microRNA
MLXIPL
MLX-interacting protein-like
mTOR
mammalian target of rapamycin
NAFLD
nonalcoholic fatty liver disease
NASH
nonalcoholic steatohepatitis
ncRNAs
non-coding RNAs
NEAT1
nuclear enriched abundant transcript 1
NTCP
sodium-taurocholate co-transporting polypeptide
PEPCK
phosphoenolpyruvate carboxykinase
PI3K
phosphatidylinositol 3-kinase
PTBP1
polypyrimidine tract-binding protein 1
SHP
small heterodimer partner
SRA
steroid receptor RNA activator
SREBP-1c
sterol regulatory element-binding protein 1
TC
total cholesterol
TG
triglyceride
ZEB1
zinc finger E-box binding homeobox 1